Differential gene expression
Malgorzata Nowicka
2024-09-30
Differential_Gene_Expression.Rmd
### Packages to compile rmarkdown
library(knitr)
library(rmarkdown)
### Packages for nice tables
library(kableExtra)
font_size <- 9
cache <- 0
### Set global option for font_size for bkable in this document
options(bkable_font_size = font_size)
options(bkable_full_width = TRUE)
knitr::opts_chunk$set(cache = cache, cache.comments = FALSE, echo = TRUE, warning = FALSE, message = FALSE, error = FALSE, fig.width = 6, fig.height = 5, fig.align = "center", tidy=TRUE, tidy.opts = list(width.cutoff = 80))
library(ggplot2)
library(SummarizedExperiment)
library(limma)
library(edgeR)
library(msigdbr)
library(BiomarkerWrappers)
ggplot2::theme_set(cowplot::theme_cowplot(12))Load RNA-seq data
gse SummarisedExperiment object contains RNA-seq data of 553 samples from the GOYA clinical trial.
The GOYA trial tested the efficacy of Gazyva (GA101) compared with Rituxan (Rituximab) in first line, untreated DLBCL patients. Patients were randomized 1:1 to either G or R combined with a CHOP chemotherapy backbone. Tumor samples were collected at baseline, RNA was isolated using RNA-Access, and RNASeq was run with TruSeq (Illumina) RNASeq.
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE125966
data(gse)
gse## class: SummarizedExperiment
## dim: 27675 553
## metadata(0):
## assays(1): counts
## rownames(27675): 1 10 ... 9994 9997
## rowData names(2): EntrezIDs Hgnc_Symbol
## colnames(553): bo21005_bo2100500001_20160904
## bo21005_bo2100500002_20160904 ... bo21005_bo2100501181_20160904
## bo21005_bo2100501182_20160901
## colData names(23): RNASeq_sample_id Run ... ipi region
colData(gse)## DataFrame with 553 rows and 23 columns
## RNASeq_sample_id Run BioProject
## <character> <character> <character>
## bo21005_bo2100500001_20160904 bo21005_bo2100500001.. SRR8512989 PRJNA518119
## bo21005_bo2100500002_20160904 bo21005_bo2100500002.. SRR8512990 PRJNA518119
## bo21005_bo2100500003_20160904 bo21005_bo2100500003.. SRR8512991 PRJNA518119
## bo21005_bo2100500004_20160827 bo21005_bo2100500004.. SRR8512992 PRJNA518119
## bo21005_bo2100500005_20160827 bo21005_bo2100500005.. SRR8512993 PRJNA518119
## ... ... ... ...
## bo21005_bo2100501175_20160911 bo21005_bo2100501175.. SRR8513393 PRJNA518119
## bo21005_bo2100501176_20160906 bo21005_bo2100501176.. SRR8513394 PRJNA518119
## bo21005_bo2100501177_20160901 bo21005_bo2100501177.. SRR8513395 PRJNA518119
## bo21005_bo2100501181_20160904 bo21005_bo2100501181.. SRR8513396 PRJNA518119
## bo21005_bo2100501182_20160901 bo21005_bo2100501182.. SRR8513397 PRJNA518119
## BioSample Experiment GEO_Accession
## <character> <character> <character>
## bo21005_bo2100500001_20160904 SAMN10853933 SRX5316605 GSM3586436
## bo21005_bo2100500002_20160904 SAMN10853932 SRX5316606 GSM3586437
## bo21005_bo2100500003_20160904 SAMN10853931 SRX5316607 GSM3586438
## bo21005_bo2100500004_20160827 SAMN10853930 SRX5316608 GSM3586439
## bo21005_bo2100500005_20160827 SAMN10853929 SRX5316609 GSM3586440
## ... ... ... ...
## bo21005_bo2100501175_20160911 SAMN10853936 SRX5317057 GSM3586984
## bo21005_bo2100501176_20160906 SAMN10853935 SRX5317058 GSM3586985
## bo21005_bo2100501177_20160901 SAMN10853934 SRX5317059 GSM3586986
## bo21005_bo2100501181_20160904 SAMN10853906 SRX5317060 GSM3586987
## bo21005_bo2100501182_20160901 SAMN10853905 SRX5317157 GSM3586988
## Instrument LibraryLayout
## <character> <character>
## bo21005_bo2100500001_20160904 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100500002_20160904 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100500003_20160904 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100500004_20160827 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100500005_20160827 Illumina HiSeq 2500 PAIRED
## ... ... ...
## bo21005_bo2100501175_20160911 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100501176_20160906 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100501177_20160901 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100501181_20160904 Illumina HiSeq 2500 PAIRED
## bo21005_bo2100501182_20160901 Illumina HiSeq 2500 PAIRED
## LibrarySelection LibrarySource MBases
## <character> <character> <integer>
## bo21005_bo2100500001_20160904 cDNA TRANSCRIPTOMIC 4621
## bo21005_bo2100500002_20160904 cDNA TRANSCRIPTOMIC 4740
## bo21005_bo2100500003_20160904 cDNA TRANSCRIPTOMIC 4504
## bo21005_bo2100500004_20160827 cDNA TRANSCRIPTOMIC 5033
## bo21005_bo2100500005_20160827 cDNA TRANSCRIPTOMIC 4906
## ... ... ... ...
## bo21005_bo2100501175_20160911 cDNA TRANSCRIPTOMIC 4695
## bo21005_bo2100501176_20160906 cDNA TRANSCRIPTOMIC 4861
## bo21005_bo2100501177_20160901 cDNA TRANSCRIPTOMIC 5486
## bo21005_bo2100501181_20160904 cDNA TRANSCRIPTOMIC 4730
## bo21005_bo2100501182_20160901 cDNA TRANSCRIPTOMIC 5295
## MBytes Platform sample_acc Sample.Name
## <integer> <character> <character> <character>
## bo21005_bo2100500001_20160904 1990 ILLUMINA SRS4313138 GSM3586436
## bo21005_bo2100500002_20160904 2066 ILLUMINA SRS4313139 GSM3586437
## bo21005_bo2100500003_20160904 1961 ILLUMINA SRS4313140 GSM3586438
## bo21005_bo2100500004_20160827 2290 ILLUMINA SRS4313141 GSM3586439
## bo21005_bo2100500005_20160827 2269 ILLUMINA SRS4313142 GSM3586440
## ... ... ... ... ...
## bo21005_bo2100501175_20160911 2563 ILLUMINA SRS4313588 GSM3586984
## bo21005_bo2100501176_20160906 2146 ILLUMINA SRS4313589 GSM3586985
## bo21005_bo2100501177_20160901 2763 ILLUMINA SRS4313591 GSM3586986
## bo21005_bo2100501181_20160904 2199 ILLUMINA SRS4313590 GSM3586987
## bo21005_bo2100501182_20160901 2498 ILLUMINA SRS4313664 GSM3586988
## source_name SRA.Study patient_id treatment
## <character> <character> <character> <character>
## bo21005_bo2100500001_20160904 DLBCL SRP183071 Patient1 Rituximab
## bo21005_bo2100500002_20160904 DLBCL SRP183071 Patient2 GA101
## bo21005_bo2100500003_20160904 DLBCL SRP183071 Patient3 GA101
## bo21005_bo2100500004_20160827 DLBCL SRP183071 Patient4 GA101
## bo21005_bo2100500005_20160827 DLBCL SRP183071 Patient5 GA101
## ... ... ... ... ...
## bo21005_bo2100501175_20160911 DLBCL SRP183071 Patient391 Rituximab
## bo21005_bo2100501176_20160906 DLBCL SRP183071 Patient392 Rituximab
## bo21005_bo2100501177_20160901 DLBCL SRP183071 Patient393 Rituximab
## bo21005_bo2100501181_20160904 DLBCL SRP183071 Patient394 Rituximab
## bo21005_bo2100501182_20160901 DLBCL SRP183071 Patient395 GA101
## chemotherapy_cycles cell_of_origin
## <integer> <character>
## bo21005_bo2100500001_20160904 6 GCB
## bo21005_bo2100500002_20160904 6 GCB
## bo21005_bo2100500003_20160904 6 GCB
## bo21005_bo2100500004_20160827 6 GCB
## bo21005_bo2100500005_20160827 6 ABC
## ... ... ...
## bo21005_bo2100501175_20160911 6 GCB
## bo21005_bo2100501176_20160906 6
## bo21005_bo2100501177_20160901 6 GCB
## bo21005_bo2100501181_20160904 8 GCB
## bo21005_bo2100501182_20160901 6 GCB
## ipi region
## <character> <character>
## bo21005_bo2100500001_20160904 High Other
## bo21005_bo2100500002_20160904 Low-Intermediate Other
## bo21005_bo2100500003_20160904 High-Intermediate Other
## bo21005_bo2100500004_20160827 High-Intermediate Other
## bo21005_bo2100500005_20160827 Low-Intermediate Other
## ... ... ...
## bo21005_bo2100501175_20160911 Low-Intermediate Western Europe
## bo21005_bo2100501176_20160906 Low-Intermediate Western Europe
## bo21005_bo2100501177_20160901 High Eastern Europe
## bo21005_bo2100501181_20160904 High-Intermediate Eastern Europe
## bo21005_bo2100501182_20160901 Low Western Europe
rowData(gse)## DataFrame with 27675 rows and 2 columns
## EntrezIDs Hgnc_Symbol
## <character> <character>
## 1 1 A1BG
## 10 10 NAT2
## 100 100 ADA
## 1000 1000 CDH2
## 10000 10000 AKT3
## ... ... ...
## 9991 9991 PTBP3
## 9992 9992 KCNE2
## 9993 9993 DGCR2
## 9994 9994 CASP8AP2
## 9997 9997 SCO2
rowData(gse)$HGNC_Symbol <- rowData(gse)$Hgnc_Symbol
## There are some duplicated Hgnc_Symbol. Replace them with EntrezIDs
table(duplicated2(rowData(gse)$HGNC_Symbol, value = TRUE))##
##
## 1777
rowData(gse)$HGNC_Symbol[is.na(rowData(gse)$HGNC_Symbol) | rowData(gse)$HGNC_Symbol %in%
""] <- rowData(gse)$EntrezIDs[is.na(rowData(gse)$HGNC_Symbol) | rowData(gse)$HGNC_Symbol %in%
""]
table(duplicated2(rowData(gse)$HGNC_Symbol, value = TRUE))## < table of extent 0 >
rowData(gse)$HGNC_Symbol[duplicated(rowData(gse)$HGNC_Symbol)] <- rowData(gse)$EntrezIDs[duplicated(rowData(gse)$HGNC_Symbol)]
table(duplicated2(rowData(gse)$HGNC_Symbol, value = TRUE))## < table of extent 0 >Load gene sets from MSigDB
For the description of the gene sets see https://www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=H
### -------------------------------------------------------------------------
### Hallmark from MSigDB
### -------------------------------------------------------------------------
geneset_df_hallmark <- msigdbr::msigdbr(species = "Homo sapiens", category = "H")
### Skip genes that are missing
table(geneset_df_hallmark$human_gene_symbol %in% rowData(gse)$HGNC_Symbol)##
## FALSE TRUE
## 108 8101
geneset_df_hallmark <- geneset_df_hallmark[geneset_df_hallmark$human_gene_symbol %in%
rowData(gse)$HGNC_Symbol, ]
### Format the gene set names
geneset_df_hallmark$gs_name2 <- gsub("_", " ", geneset_df_hallmark$gs_name)
geneset_df_hallmark$gs_name2 <- gsub("^HALLMARK ", "", geneset_df_hallmark$gs_name2)
geneset_list_hallmark <- split(geneset_df_hallmark$human_gene_symbol, geneset_df_hallmark$gs_name2)
geneset_extra_hallmark <- data.frame(GenesetID = names(geneset_list_hallmark), Category = "HALLMARK",
stringsAsFactors = FALSE)Differential gene expression between cell of origin
table(gse$cell_of_origin)##
## ABC GCB UNCLASSIFIED
## 15 151 298 89
gse$DE_var <- factor(gse$cell_of_origin, levels = c("GCB", "UNCLASSIFIED", "ABC"))
### Remove samples with NAs
gse_dge <- gse[, !is.na(gse$DE_var)]
expr <- assay(gse_dge)
pdata <- data.frame(colData(gse_dge))
fdata <- data.frame(rowData(gse_dge))
### Number of samples
x <- table(pdata$DE_var)
rownames(x) <- gsub("_", " ", rownames(x))
kable(x, caption = "Number of samples used for the analysis.", booktabs = TRUE, linesep = "") %>%
kable_styling(bootstrap_options = c("condensed", "bordered", "striped"), latex_options = c("HOLD_position",
"striped"), full_width = FALSE, font_size = font_size)| Var1 | Freq |
|---|---|
| GCB | 298 |
| UNCLASSIFIED | 89 |
| ABC | 151 |
Define the comparisons of interest
# Design matrix. We include in the model information about treatment arm and
# time point
design <- model.matrix(~0 + DE_var, data = pdata)
colnames(design)
# Generate contrast of interest - comparisons that we want to test Group names
# must exactly match the column names of the design matrix.
contrast_matrix <- makeContrasts(UNCLASSIFIED_vs_GCB = DE_varUNCLASSIFIED - DE_varGCB,
ABC_vs_GCB = DE_varABC - DE_varGCB, levels = design)
contrasts <- colnames(contrast_matrix)
group_var = "DE_var"
contrast_mapping = c(UNCLASSIFIED_vs_GCB = "UNCLASSIFIED", ABC_vs_GCB = "ABC")
pval <- 0.05Run limma voom
### ---------------------------------------------------------------------- DE
### analysis using voom and limma model approach
### ----------------------------------------------------------------------
# Obtain CPMs
myCPM <- edgeR::cpm(expr)
# We would like to skip genes that have very low expression. We keep genes
# that have expression higher than 0.5 CPM in at least X samples, which is the
# size of the smallest subgroup that we want to compare
genes2keep <- rowSums(myCPM > 0.5) >= min(table(pdata[, group_var]))
table(genes2keep)## genes2keep
## FALSE TRUE
## 11591 16084
# Subset the rows of expr to keep the more highly expressed genes
y <- DGEList(expr[genes2keep, ], genes = fdata[genes2keep, ])
# Apply normalization to DGEList object
y <- calcNormFactors(y)
# Estimate voom mean-variance trend and precision weights
v <- voom(y, design = design, plot = TRUE)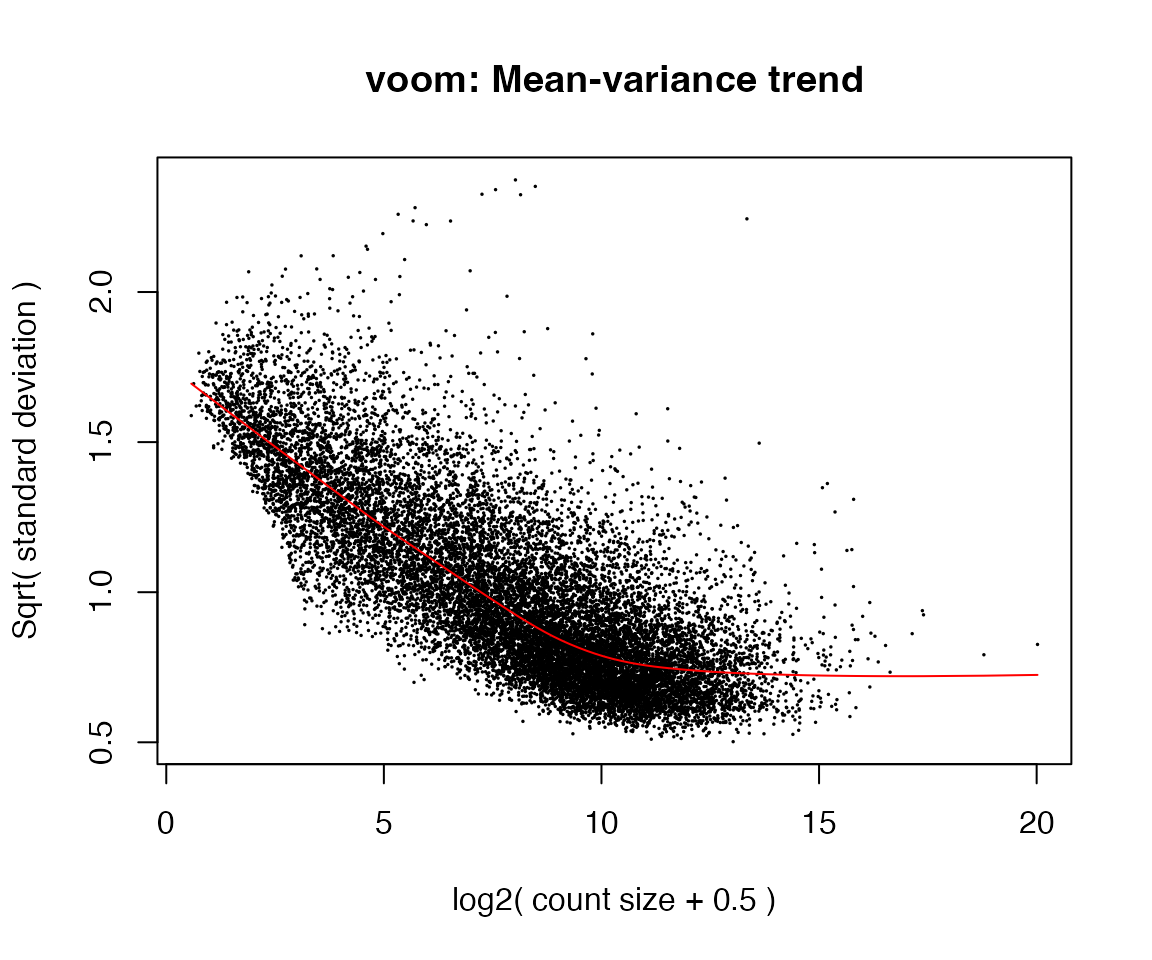
# Estimate linear model
fit <- lmFit(v, design = design)
# We apply the contrasts matrix to the fit object to get the statistics and
# estimated parameters of our comparison that we are interested in
fit <- contrasts.fit(fit, contrasts = contrast_matrix)
# The final step is to call the eBayes function, which performs empirical Bayes
# shrinkage on the variances, and estimates moderated t-statistics and the
# associated p-values
fit <- eBayes(fit)
# -------------------------------------------------------------------------
# Tables with number of DE genes
# -------------------------------------------------------------------------
summa_fit_lfc0 <- decideTests(fit, p.value = pval, lfc = 0)
x <- t(as.data.frame.matrix(summary(summa_fit_lfc0)))
kable(x, caption = paste0("Number of DE genes (adj. p-value < ", pval, ", |LogFC| > 0)."),
booktabs = TRUE, linesep = "") %>%
kable_styling(bootstrap_options = c("condensed", "bordered", "striped"), latex_options = c("HOLD_position",
"striped"), full_width = FALSE, font_size = font_size) %>%
print()| Down | NotSig | Up | |
|---|---|---|---|
| UNCLASSIFIED_vs_GCB | 1010 | 14114 | 960 |
| ABC_vs_GCB | 4607 | 7034 | 4443 |
summa_fit_lfc1 <- decideTests(fit, p.value = pval, lfc = 1)
x <- t(as.data.frame.matrix(summary(summa_fit_lfc1)))
kable(x, caption = paste0("Number of DE genes (adj. p-value < ", pval, ", |LogFC| > 1)."),
booktabs = TRUE, linesep = "") %>%
kable_styling(bootstrap_options = c("condensed", "bordered", "striped"), latex_options = c("HOLD_position",
"striped"), full_width = FALSE, font_size = font_size) %>%
print()| Down | NotSig | Up | |
|---|---|---|---|
| UNCLASSIFIED_vs_GCB | 120 | 15872 | 92 |
| ABC_vs_GCB | 691 | 15156 | 237 |
# -------------------------------------------------------------------------
# plotMD
# -------------------------------------------------------------------------
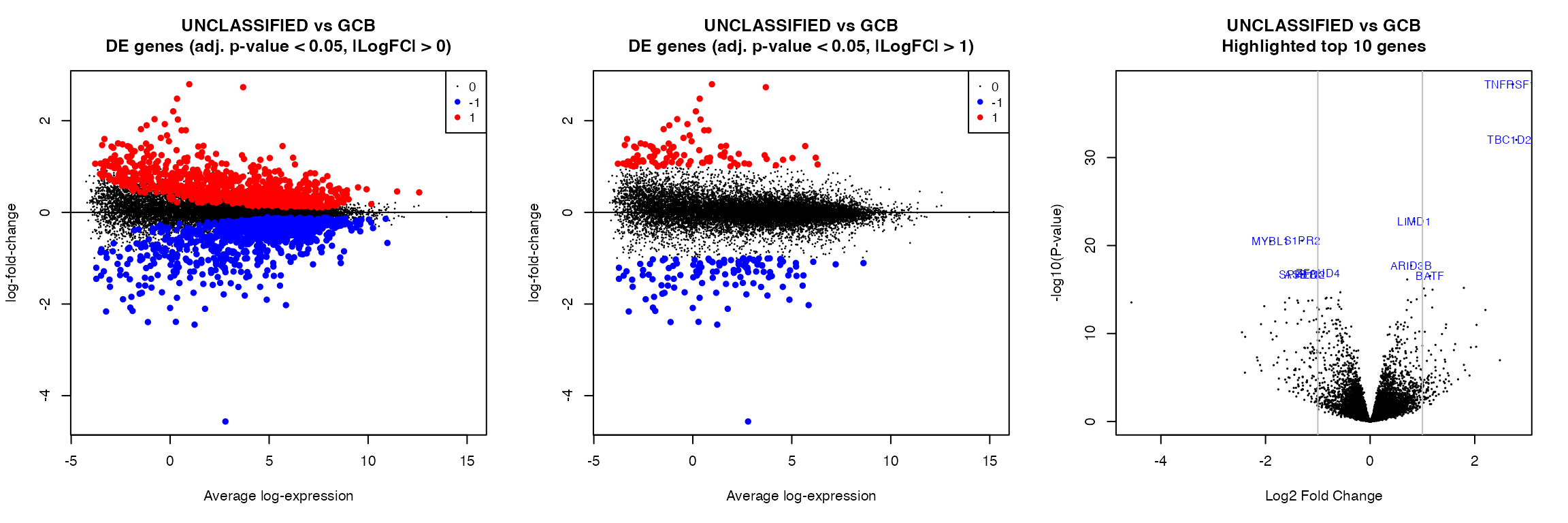
par(mfrow = c(1, 3))
for (k in 1:length(colnames(contrast_matrix))) {
# k = 1
contrast <- colnames(contrast_matrix)[k]
plotMD(fit, coef = contrast, status = summa_fit_lfc0[, contrast], values = c(-1,
1), hl.col = c("blue", "red"), main = paste0(gsub("_", " ", contrast), "\nDE genes (adj. p-value < ",
pval, ", |LogFC| > 0)"), cex.main = 1.2)
abline(h = 0)
plotMD(fit, coef = contrast, status = summa_fit_lfc1[, contrast], values = c(-1,
1), hl.col = c("blue", "red"), main = paste0(gsub("_", " ", contrast), "\nDE genes (adj. p-value < ",
pval, ", |LogFC| > 1)"), cex.main = 1.2)
abline(h = 0)
volcanoplot(fit, coef = contrast, highlight = 10, names = fit$genes$HGNC_Symbol,
main = paste0(gsub("_", " ", contrast), "\nHighlighted top 10 genes"))
abline(v = c(-1, 1), col = "grey")
}
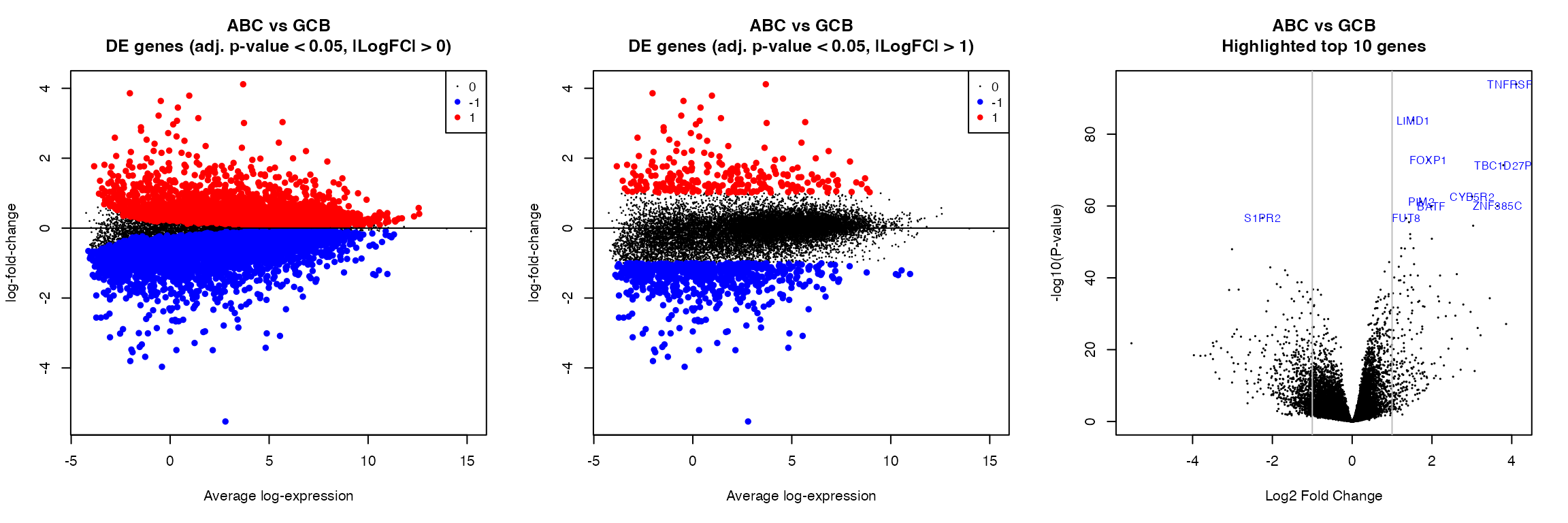
# -------------------------------------------------------------------------
# topTable
# -------------------------------------------------------------------------
topTable <- wrapper_merge_topTables(fit = fit, contrasts = colnames(contrast_matrix),
gene_vars = c("EntrezIDs", "HGNC_Symbol"), pval = pval)
rownames(topTable) <- topTable$HGNC_Symbol
head(topTable)## EntrezIDs HGNC_Symbol logFC_UNCLASSIFIED_vs_GCB
## A1BG 1 A1BG 0.05258256
## ADA 100 ADA 0.04592907
## CDH2 1000 CDH2 -0.09764943
## AKT3 10000 AKT3 -0.03206742
## MED6 10001 MED6 -0.07466933
## NAALAD2 10003 NAALAD2 0.03534835
## AveExpr_UNCLASSIFIED_vs_GCB t_UNCLASSIFIED_vs_GCB
## A1BG 1.183651 0.3896911
## ADA 6.836000 0.3804623
## CDH2 2.816823 -0.4727322
## AKT3 5.454880 -0.3258116
## MED6 4.797126 -1.3328362
## NAALAD2 -1.094422 0.1547457
## P.Value_UNCLASSIFIED_vs_GCB adj.P.Val_UNCLASSIFIED_vs_GCB
## A1BG 0.6969187 0.8550797
## ADA 0.7037519 0.8587647
## CDH2 0.6365953 0.8204983
## AKT3 0.7446931 0.8813318
## MED6 0.1831478 0.4264694
## NAALAD2 0.8770796 0.9462036
## sign.P.Val_UNCLASSIFIED_vs_GCB summary_pval5_lfc0_UNCLASSIFIED_vs_GCB
## A1BG 0.15681790 0
## ADA 0.15258040 0
## CDH2 -0.19613659 0
## AKT3 -0.12802266 0
## MED6 -0.73719829 0
## NAALAD2 0.05696097 0
## summary_pval5_lfc1_UNCLASSIFIED_vs_GCB logFC_ABC_vs_GCB
## A1BG 0 0.08738254
## ADA 0 0.26680147
## CDH2 0 -0.40113964
## AKT3 0 -0.22395773
## MED6 0 0.08464979
## NAALAD2 0 -0.49081115
## AveExpr_ABC_vs_GCB t_ABC_vs_GCB P.Value_ABC_vs_GCB adj.P.Val_ABC_vs_GCB
## A1BG 1.183651 0.7778443 0.437001537 0.51700866
## ADA 6.836000 2.6816020 0.007551309 0.01575295
## CDH2 2.816823 -2.2295012 0.026191155 0.04690553
## AKT3 5.454880 -2.7059657 0.007025991 0.01478943
## MED6 4.797126 1.8450846 0.065572741 0.10359218
## NAALAD2 -1.094422 -2.4368811 0.015136556 0.02908331
## sign.P.Val_ABC_vs_GCB summary_pval5_lfc0_ABC_vs_GCB
## A1BG 0.359517 0
## ADA 2.121978 1
## CDH2 -1.581845 -1
## AKT3 -2.153292 -1
## MED6 1.183277 0
## NAALAD2 -1.819973 -1
## summary_pval5_lfc1_ABC_vs_GCB
## A1BG 0
## ADA 0
## CDH2 0
## AKT3 0
## MED6 0
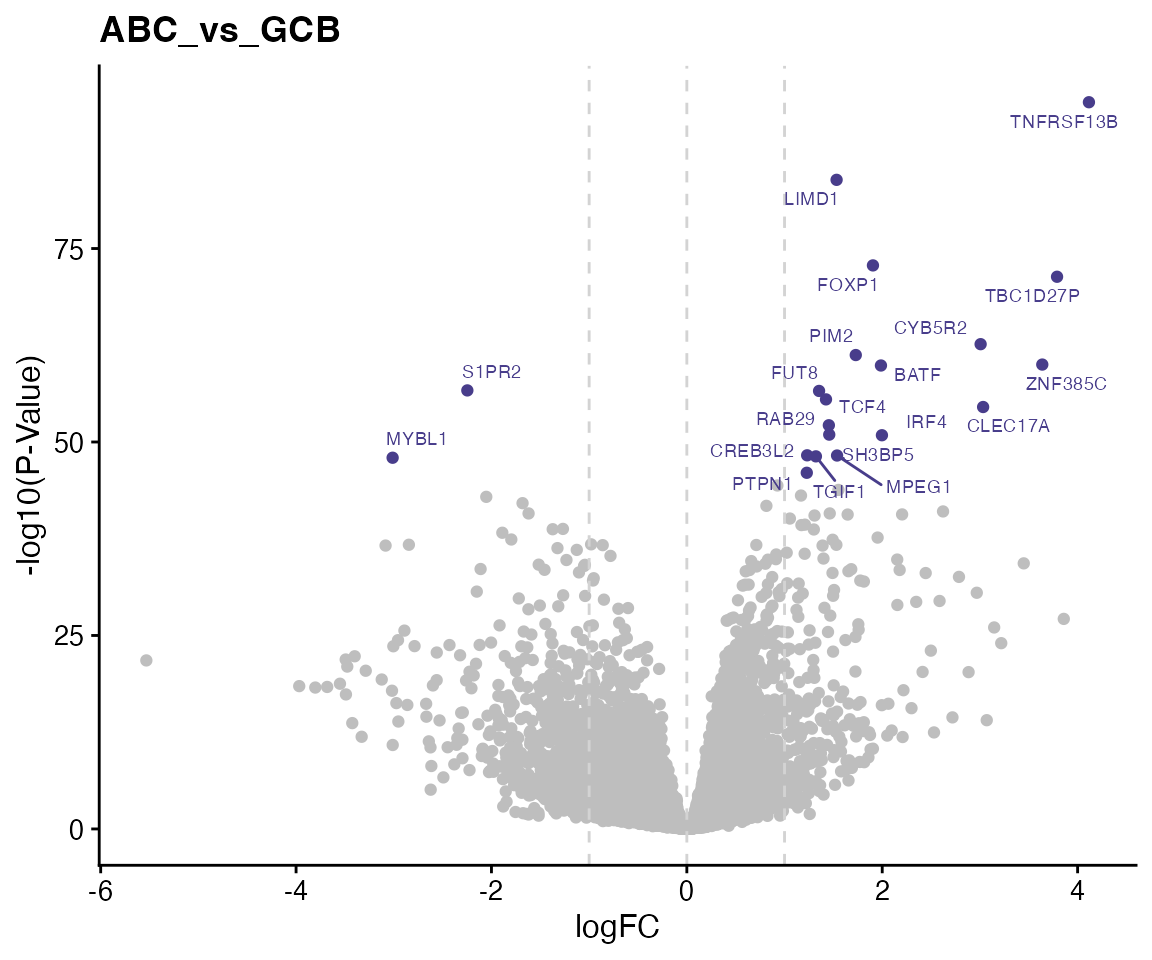
## NAALAD2 0Volcano plots
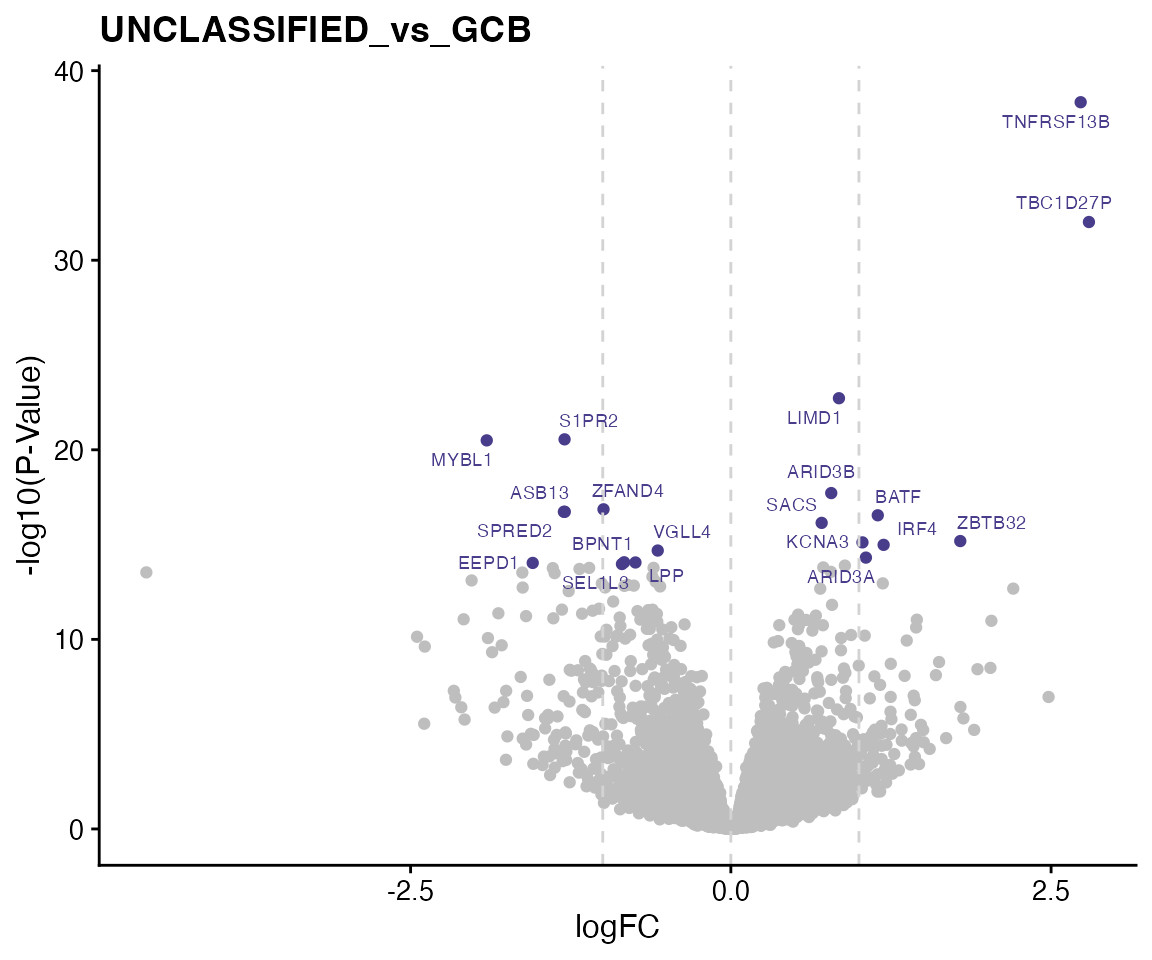
We highlight top 20 genes in the volcano plot.
for (i in 1:length(contrasts)) {
# i = 1
contrast <- contrasts[i]
direction <- "both"
topTable_both <- wrapper_dispaly_significant_genes(topTable, contrast = contrast,
direction = direction, topn = 20, pval = 1, lfc = 0, gene_vars = c("EntrezIDs",
"HGNC_Symbol"), lfc_prefix = "logFC", pval_prefix = "P.Value", adjp_prefix = "adj.P.Val")
ggdata <- topTable[, c("HGNC_Symbol", paste0(c("logFC_", "P.Value_"), contrast))]
colnames(ggdata) <- c("HGNC_Symbol", "logFC", "P.Value")
ggdata$P.Value <- -log10(ggdata$P.Value)
ggdata$Label <- ggdata$HGNC_Symbol
ggdata$Label[!ggdata$Label %in% bresults(topTable_both)$HGNC_Symbol] <- NA
ggdata$Highlight <- is.na(ggdata$Label)
ggp <- ggplot(ggdata, aes(x = logFC, y = P.Value, color = Highlight, label = Label)) +
geom_point() + geom_vline(xintercept = 0, linetype = 2, color = "lightgrey") +
geom_vline(xintercept = c(-1, 1), linetype = 2, color = "lightgrey") + ggrepel::geom_text_repel(size = 2.5) +
labs(title = contrast) + ylab("-log10(P-Value)") + theme(legend.position = "none") +
scale_color_manual(values = c("darkslateblue", "grey"))
print(ggp)
}
Correlation between LogFC
x <- topTable[, paste0("logFC_", contrasts), drop = FALSE]
colnames(x) <- gsub("_", " ", colnames(x))
correlation <- cor(x, method = "spearman", use = "pairwise.complete.obs")
### Corrplot
corrplot::corrplot(correlation, method = "color", addCoef.col = "black", number.digits = 1,
number.cex = 0.75, order = "hclust", hclust.method = "complete", type = "full",
tl.col = "black", tl.cex = 1, cl.pos = "n")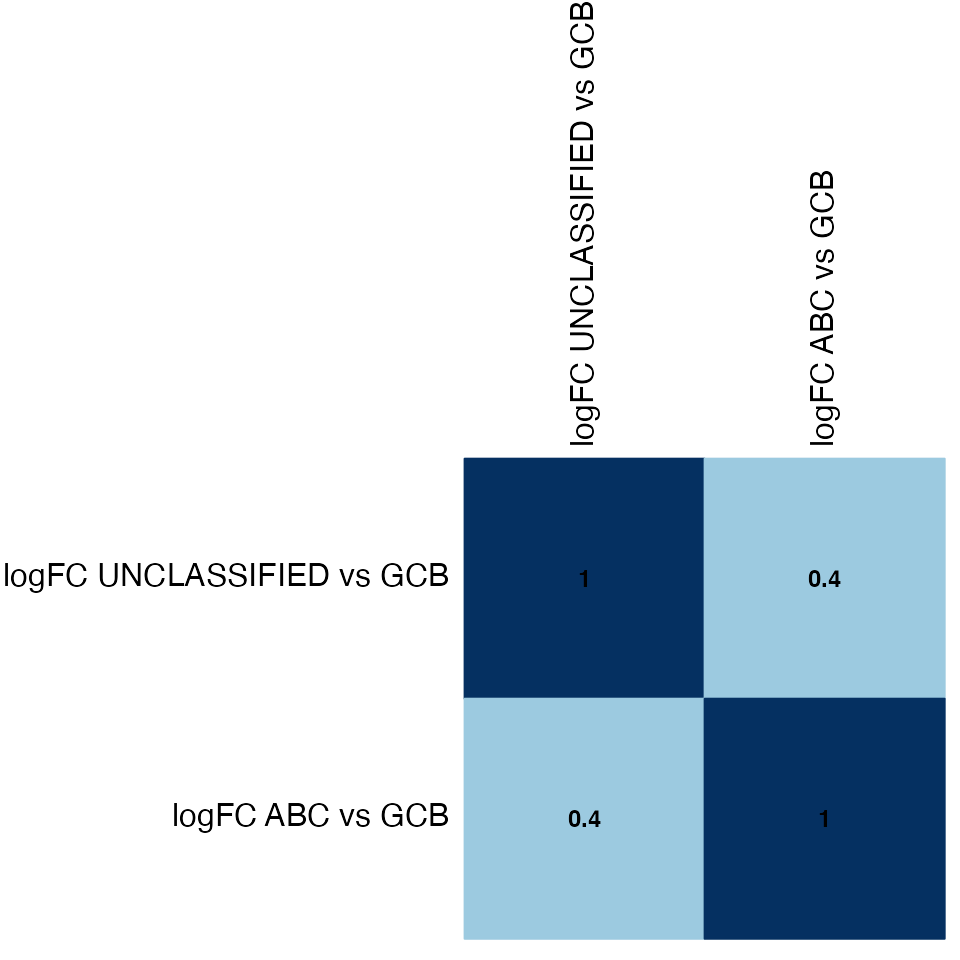
### Scatter plot with LogFC
ggdf <- topTable
logFC1 <- paste0("logFC_", contrasts[1])
logFC2 <- paste0("logFC_", contrasts[2])
logFC_cor <- round(cor(ggdf[, logFC1], ggdf[, logFC2], method = "spearman", use = "pairwise.complete.obs"),
2)
logfc_range <- range(c(ggdf[, logFC1], ggdf[, logFC2]))
wrapper_point_plot_core(ggdf, x_var = logFC1, y_var = logFC2, title = paste0("Spearman's R = ",
logFC_cor), point_shape = 20, background_grid_major = "xy", xlim = logfc_range,
ylim = logfc_range) + geom_abline(slope = 1, intercept = 0, color = "grey")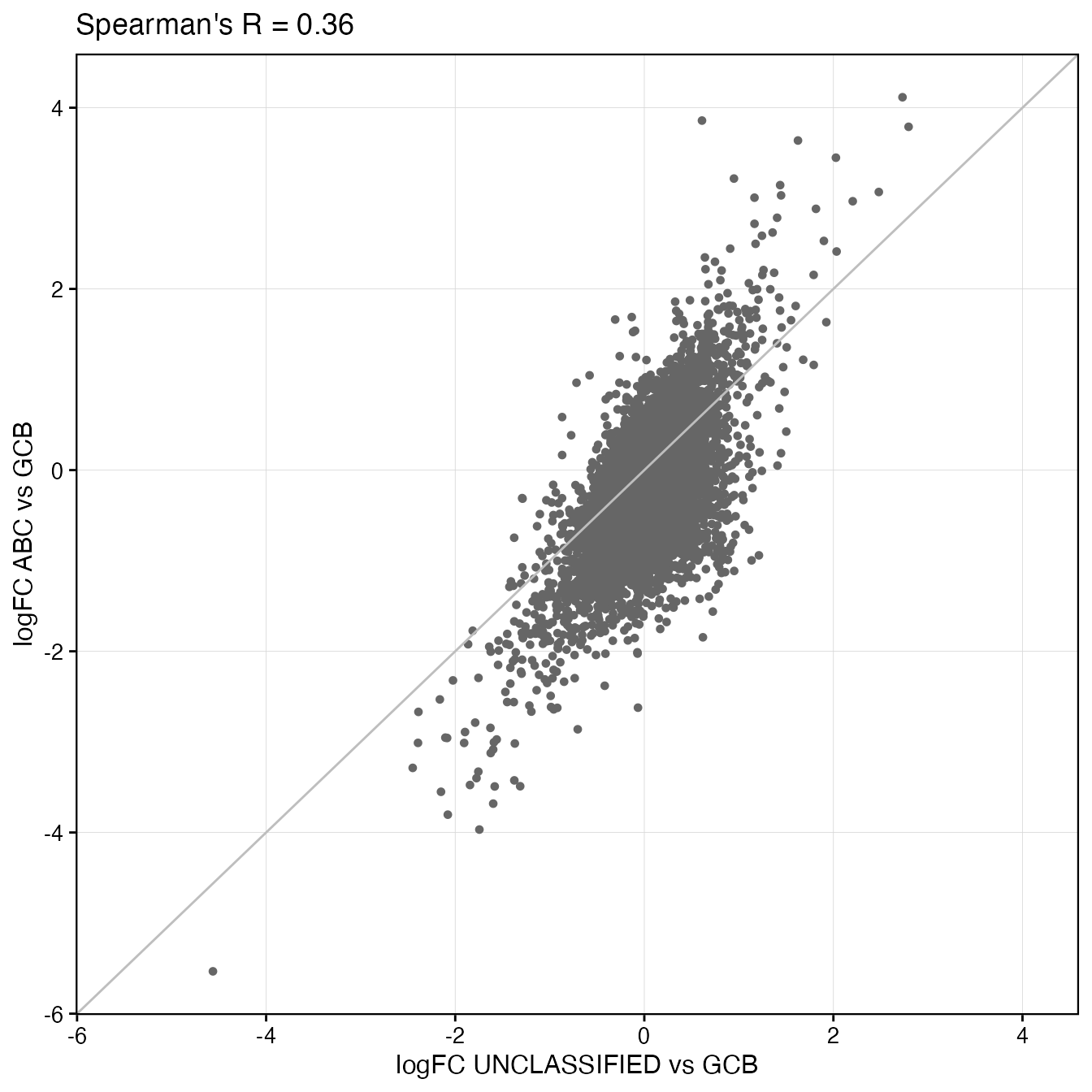
### Venn diagram
set1 <- contrasts[1]
set2 <- contrasts[2]
venn_data <- topTable[, paste0("summary_pval5_lfc0_", c(set1, set2))]
colnames(venn_data) <- c(set1, set2)
vennDiagram(venn_data, include = c("up", "down"), counts.col = c("red", "blue"),
circle.col = c("blue", "orange", "grey"), main = "|LogFC| > 0, adj. p-val < 0.05",
cex = c(0.8, 0.6, 0.6))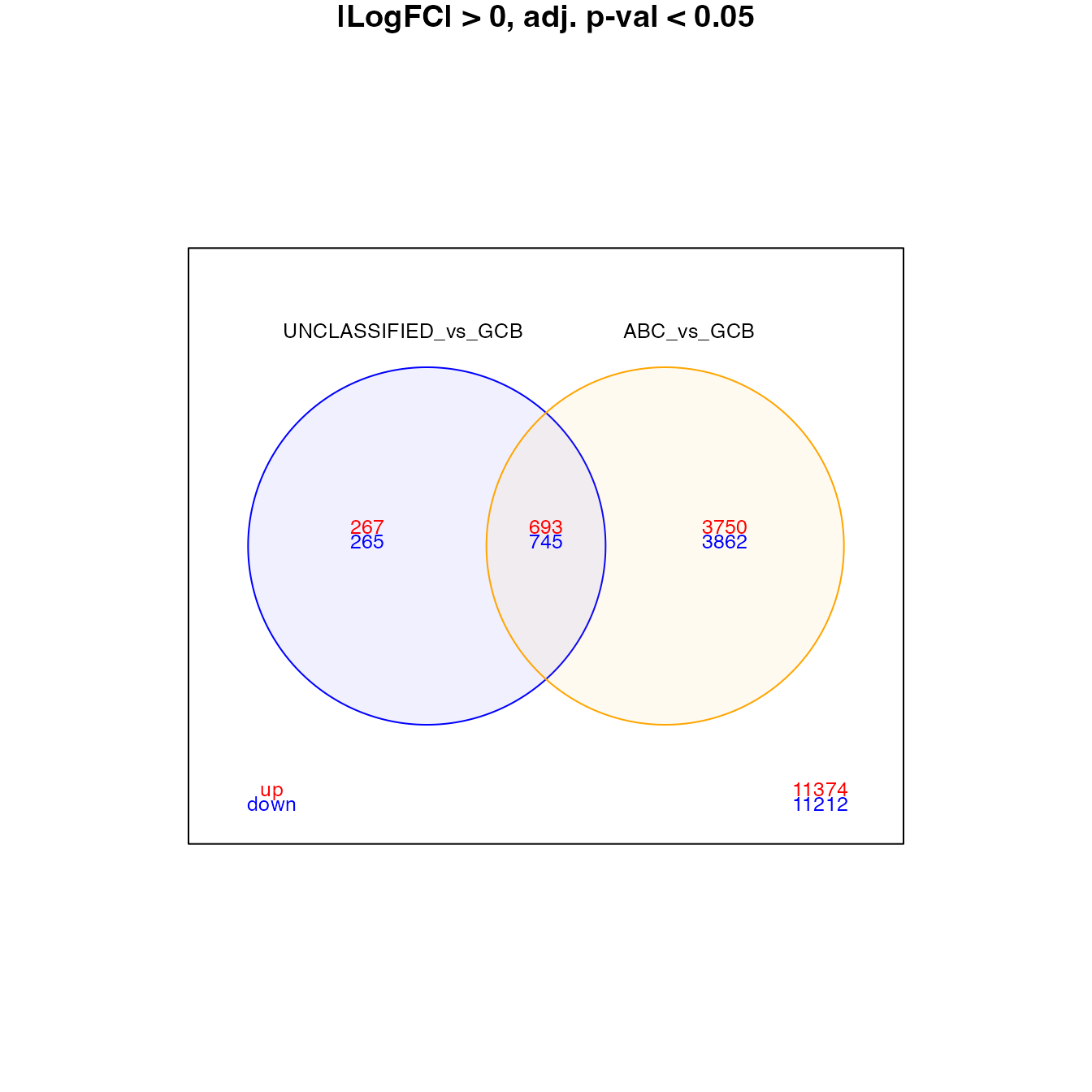
venn_data <- topTable[, paste0("summary_pval5_lfc1_", c(set1, set2))]
colnames(venn_data) <- c(set1, set2)
vennDiagram(venn_data, include = c("up", "down"), counts.col = c("red", "blue"),
circle.col = c("blue", "orange", "grey"), main = "|LogFC| > 1, adj. p-val < 0.05",
cex = c(0.8, 0.6, 0.6))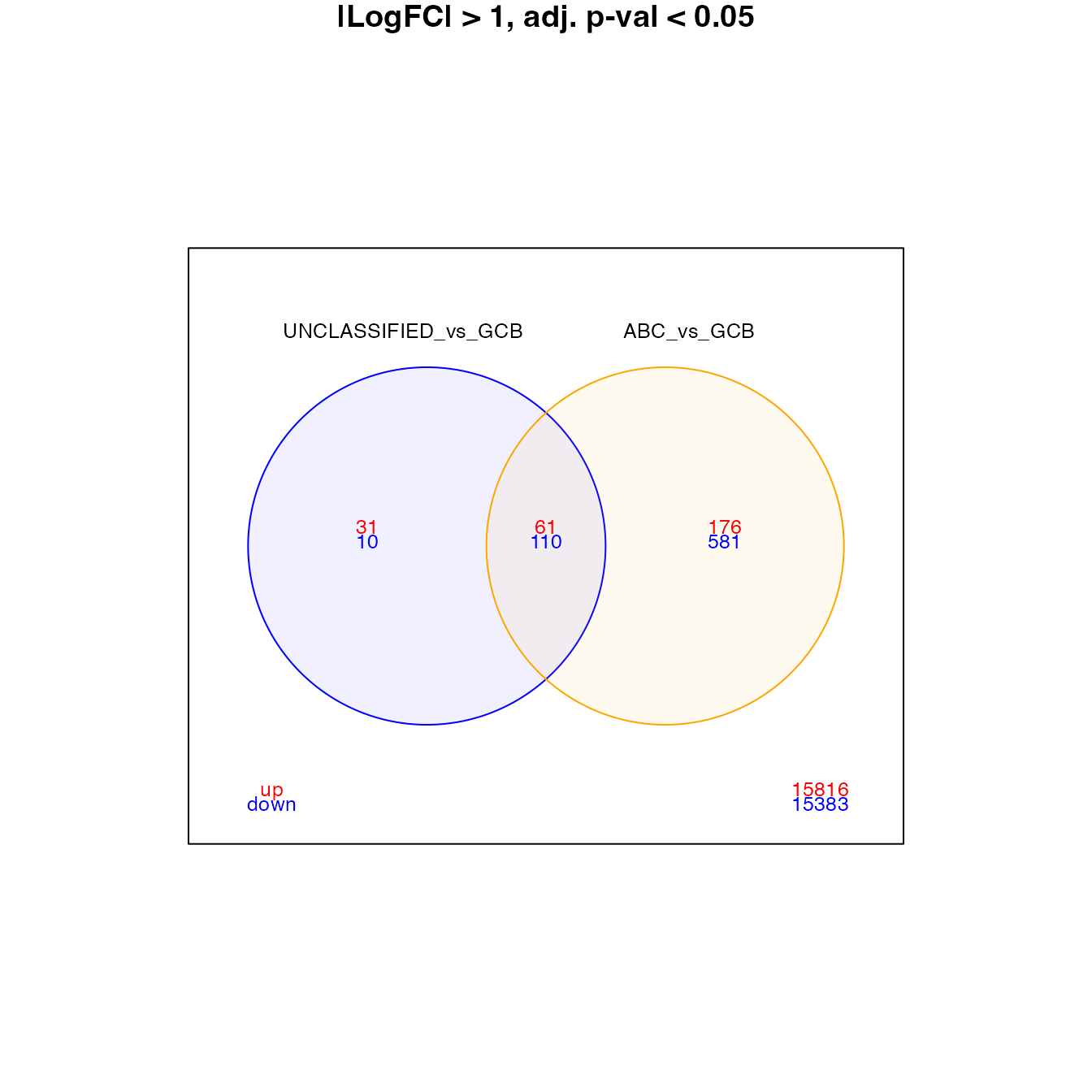
Significant results
# ------------------------------ Generate tables with significant results
# ------------------------------
significant_genes_list <- lapply(1:length(contrasts), function(i) {
# i = 5
contrast <- contrasts[i]
cat(paste0("\n\n### Testing for ", gsub("_", " ", contrast), "\n\n"))
direction <- "up"
topTable_up <- wrapper_dispaly_significant_genes(topTable, contrast = contrast,
direction = direction, topn = 30, pval = pval, lfc = 0, gene_vars = c("EntrezIDs",
"HGNC_Symbol"), lfc_prefix = "logFC", pval_prefix = "P.Value", adjp_prefix = "adj.P.Val")
print(topTable_up)
direction <- "down"
topTable_down <- wrapper_dispaly_significant_genes(topTable, contrast = contrast,
direction = direction, topn = 30, pval = pval, lfc = 0, gene_vars = c("EntrezIDs",
"HGNC_Symbol"), lfc_prefix = "logFC", pval_prefix = "P.Value", adjp_prefix = "adj.P.Val")
print(topTable_down)
significant_genes <- c(bresults(topTable_up)$HGNC_Symbol, bresults(topTable_down)$HGNC_Symbol)
})Testing for UNCLASSIFIED vs GCB
| EntrezIDs | HGNC_Symbol | logFC | P.Value | adj.P.Val |
|---|---|---|---|---|
| 23495 | TNFRSF13B | 2.73 | <1e-38 *** | <1e-34 *** |
| 96597 | TBC1D27P | 2.80 | <1e-32 *** | <1e-28 *** |
| 8994 | LIMD1 | 0.84 | <1e-22 *** | <1e-18 *** |
| 10620 | ARID3B | 0.78 | <1e-17 *** | <1e-14 *** |
| 10538 | BATF | 1.15 | <1e-16 *** | <1e-13 *** |
| 26278 | SACS | 0.71 | <1e-16 *** | <1e-12 *** |
| 27033 | ZBTB32 | 1.79 | <1e-15 *** | <1e-12 *** |
| 3738 | KCNA3 | 1.03 | <1e-15 *** | <1e-12 *** |
| 3662 | IRF4 | 1.19 | <1e-14 *** | <1e-11 *** |
| 1820 | ARID3A | 1.05 | <1e-14 *** | <1e-11 *** |
| 11040 | PIM2 | 0.89 | <1e-13 *** | <1e-11 *** |
| 2530 | FUT8 | 0.72 | <1e-13 *** | <1e-10 *** |
| 375387 | NRROS | 0.78 | <1e-13 *** | <1e-10 *** |
| 9767 | JADE3 | 1.19 | <1e-12 *** | <1e-10 *** |
| 128853 | DUSP15 | 2.20 | <1e-12 *** | <1e-10 *** |
| 5142 | PDE4B | 0.70 | <1e-12 *** | <1e-10 *** |
| 27086 | FOXP1 | 0.79 | <1e-11 *** | <1e-09 *** |
| 254531 | LPCAT4 | 0.53 | <1e-11 *** | <1e-08 *** |
| 5770 | PTPN1 | 0.66 | <1e-11 *** | <1e-08 *** |
| 10382 | TUBB4A | 1.45 | <1e-11 *** | <1e-08 *** |
| 10625 | IVNS1ABP | 0.50 | <1e-11 *** | <1e-08 *** |
| 84166 | NLRC5 | 0.59 | <1e-11 *** | <1e-08 *** |
| 79152 | FA2H | 2.03 | <1e-10 *** | <1e-08 *** |
| 152137 | CCDC50 | 0.64 | <1e-10 *** | <1e-08 *** |
| 9575 | CLOCK | 0.38 | <1e-10 *** | <1e-08 *** |
| 8934 | RAB29 | 0.72 | <1e-10 *** | <1e-08 *** |
| 64784 | CRTC3 | 0.53 | <1e-10 *** | <1e-08 *** |
| 388512 | CLEC17A | 1.45 | <1e-10 *** | <1e-08 *** |
| 23303 | KIF13B | 0.52 | <1e-10 *** | <1e-08 *** |
| 953 | ENTPD1 | 0.64 | <1e-10 *** | <1e-08 *** |
| EntrezIDs | HGNC_Symbol | logFC | P.Value | adj.P.Val |
|---|---|---|---|---|
| 9294 | S1PR2 | -1.30 | <1e-20 *** | <1e-16 *** |
| 4603 | MYBL1 | -1.91 | <1e-20 *** | <1e-16 *** |
| 93550 | ZFAND4 | -0.99 | <1e-16 *** | <1e-13 *** |
| 79754 | ASB13 | -1.30 | <1e-16 *** | <1e-13 *** |
| 200734 | SPRED2 | -1.30 | <1e-16 *** | <1e-13 *** |
| 9686 | VGLL4 | -0.57 | <1e-14 *** | <1e-11 *** |
| 10380 | BPNT1 | -0.83 | <1e-14 *** | <1e-11 *** |
| 4026 | LPP | -0.74 | <1e-14 *** | <1e-11 *** |
| 80820 | EEPD1 | -1.55 | <1e-14 *** | <1e-11 *** |
| 23231 | SEL1L3 | -0.85 | <1e-13 *** | <1e-11 *** |
| 3065 | HDAC1 | -0.60 | <1e-13 *** | <1e-10 *** |
| 4033 | LRMP | -1.11 | <1e-13 *** | <1e-10 *** |
| 55534 | MAML3 | -1.39 | <1e-13 *** | <1e-10 *** |
| 56941 | HMCES | -1.18 | <1e-13 *** | <1e-10 *** |
| 327657 | SERPINA9 | -4.56 | <1e-13 *** | <1e-10 *** |
| 440352 | SNX29P2 | -1.63 | <1e-13 *** | <1e-10 *** |
| 23189 | KANK1 | -1.38 | <1e-13 *** | <1e-10 *** |
| 54906 | FAM208B | -0.61 | <1e-13 *** | <1e-10 *** |
| 2334 | AFF2 | -2.02 | <1e-13 *** | <1e-10 *** |
| 88455 | ANKRD13A | -0.59 | <1e-13 *** | <1e-10 *** |
| 65108 | MARCKSL1 | -1.01 | <1e-12 *** | <1e-10 *** |
| 604 | BCL6 | -0.80 | <1e-12 *** | <1e-10 *** |
| 23180 | RFTN1 | -0.76 | <1e-12 *** | <1e-10 *** |
| 10783 | NEK6 | -0.83 | <1e-12 *** | <1e-10 *** |
| 54431 | DNAJC10 | -0.55 | <1e-12 *** | <1e-10 *** |
| 5218 | CDK14 | -0.98 | <1e-12 *** | <1e-10 *** |
| 10734 | STAG3 | -1.63 | <1e-12 *** | <1e-10 *** |
| 26228 | STAP1 | -1.27 | <1e-12 *** | <1e-09 *** |
| 100129034 | LOC100129034 | -0.92 | <1e-11 *** | <1e-09 *** |
| 23635 | SSBP2 | -1.03 | <1e-11 *** | <1e-09 *** |
Testing for ABC vs GCB
| EntrezIDs | HGNC_Symbol | logFC | P.Value | adj.P.Val |
|---|---|---|---|---|
| 23495 | TNFRSF13B | 4.12 | <1e-93 *** | <1e-89 *** |
| 8994 | LIMD1 | 1.53 | <1e-83 *** | <1e-79 *** |
| 27086 | FOXP1 | 1.91 | <1e-72 *** | <1e-69 *** |
| 96597 | TBC1D27P | 3.79 | <1e-71 *** | <1e-67 *** |
| 51700 | CYB5R2 | 3.01 | <1e-62 *** | <1e-59 *** |
| 11040 | PIM2 | 1.73 | <1e-61 *** | <1e-57 *** |
| 201181 | ZNF385C | 3.64 | <1e-60 *** | <1e-56 *** |
| 10538 | BATF | 1.99 | <1e-59 *** | <1e-56 *** |
| 2530 | FUT8 | 1.35 | <1e-56 *** | <1e-53 *** |
| 6925 | TCF4 | 1.42 | <1e-55 *** | <1e-52 *** |
| 388512 | CLEC17A | 3.03 | <1e-54 *** | <1e-51 *** |
| 8934 | RAB29 | 1.45 | <1e-52 *** | <1e-49 *** |
| 9467 | SH3BP5 | 1.46 | <1e-50 *** | <1e-47 *** |
| 3662 | IRF4 | 2.00 | <1e-50 *** | <1e-47 *** |
| 64764 | CREB3L2 | 1.23 | <1e-48 *** | <1e-45 *** |
| 219972 | MPEG1 | 1.54 | <1e-48 *** | <1e-45 *** |
| 7050 | TGIF1 | 1.32 | <1e-48 *** | <1e-45 *** |
| 5770 | PTPN1 | 1.23 | <1e-46 *** | <1e-43 *** |
| 79718 | TBL1XR1 | 0.93 | <1e-44 *** | <1e-41 *** |
| 3738 | KCNA3 | 1.55 | <1e-43 *** | <1e-40 *** |
| 152137 | CCDC50 | 1.17 | <1e-43 *** | <1e-40 *** |
| 4067 | LYN | 0.81 | <1e-41 *** | <1e-38 *** |
| 54836 | BSPRY | 2.62 | <1e-41 *** | <1e-38 *** |
| 6689 | SPIB | 1.46 | <1e-40 *** | <1e-38 *** |
| 80183 | RUBCNL | 2.20 | <1e-40 *** | <1e-37 *** |
| 894 | CCND2 | 1.65 | <1e-40 *** | <1e-37 *** |
| 29760 | BLNK | 1.31 | <1e-40 *** | <1e-37 *** |
| 55114 | ARHGAP17 | 1.05 | <1e-40 *** | <1e-37 *** |
| 90427 | BMF | 1.21 | <1e-39 *** | <1e-36 *** |
| 375387 | NRROS | 1.17 | <1e-39 *** | <1e-36 *** |
| EntrezIDs | HGNC_Symbol | logFC | P.Value | adj.P.Val |
|---|---|---|---|---|
| 9294 | S1PR2 | -2.25 | <1e-56 *** | <1e-53 *** |
| 4603 | MYBL1 | -3.01 | <1e-47 *** | <1e-45 *** |
| 26053 | AUTS2 | -2.05 | <1e-42 *** | <1e-40 *** |
| 5218 | CDK14 | -1.68 | <1e-42 *** | <1e-39 *** |
| 22898 | DENND3 | -1.62 | <1e-40 *** | <1e-38 *** |
| 23231 | SEL1L3 | -1.27 | <1e-38 *** | <1e-36 *** |
| 10783 | NEK6 | -1.37 | <1e-38 *** | <1e-36 *** |
| 23635 | SSBP2 | -1.89 | <1e-38 *** | <1e-35 *** |
| 79754 | ASB13 | -1.80 | <1e-37 *** | <1e-34 *** |
| 24149 | ZNF318 | -0.98 | <1e-36 *** | <1e-34 *** |
| 440352 | SNX29P2 | -2.85 | <1e-36 *** | <1e-34 *** |
| 29994 | BAZ2B | -0.86 | <1e-36 *** | <1e-34 *** |
| 4311 | MME | -3.08 | <1e-36 *** | <1e-34 *** |
| 93550 | ZFAND4 | -1.32 | <1e-36 *** | <1e-33 *** |
| 3707 | ITPKB | -1.13 | <1e-36 *** | <1e-33 *** |
| 50807 | ASAP1 | -0.78 | <1e-35 *** | <1e-32 *** |
| 23648 | SSBP3 | -1.23 | <1e-34 *** | <1e-32 *** |
| 4052 | LTBP1 | -1.52 | <1e-34 *** | <1e-31 *** |
| 5747 | PTK2 | -1.04 | <1e-34 *** | <1e-31 *** |
| 7414 | VCL | -1.05 | <1e-33 *** | <1e-31 *** |
| 55534 | MAML3 | -2.11 | <1e-33 *** | <1e-31 *** |
| 100129034 | LOC100129034 | -1.46 | <1e-33 *** | <1e-31 *** |
| 55824 | PAG1 | -1.10 | <1e-33 *** | <1e-30 *** |
| 25932 | CLIC4 | -0.95 | <1e-32 *** | <1e-30 *** |
| 8895 | CPNE3 | -0.96 | <1e-32 *** | <1e-29 *** |
| 80820 | EEPD1 | -2.15 | <1e-30 *** | <1e-28 *** |
| 5098 | PCDHGC3 | -1.27 | <1e-30 *** | <1e-27 *** |
| 79666 | PLEKHF2 | -1.04 | <1e-30 *** | <1e-27 *** |
| 26230 | TIAM2 | -1.72 | <1e-29 *** | <1e-27 *** |
| 23139 | MAST2 | -0.85 | <1e-29 *** | <1e-27 *** |
names(significant_genes_list) <- contrastsSummary
significant_genes <- unique(unlist(significant_genes_list))
topTable_significant_genes <- topTable[significant_genes, , drop = FALSE]
# -----------------------------------------------------------------------
# Clustering by LogFC
# -----------------------------------------------------------------------
data_lfc <- wrapper_extract_from_topTable(topTable_significant_genes, extract_prefix = "logFC")
if (ncol(data_lfc) > 1) {
# Cluster rows using hierarchical clustering
d <- dist(data_lfc, method = "euclidean")
cluster_rows <- hclust(d, method = "complete")
clustering_order <- cluster_rows$order
} else {
cluster_rows <- FALSE
clustering_order <- 1:nrow(data_lfc)
}
### logFC_dotplot
wrapper_logFC_dotplot(topTable_significant_genes[clustering_order, ], gene_var = "HGNC_Symbol",
lfc_prefix = "logFC", pval_prefix = "P.Value", adjp_prefix = "adj.P.Val", pval = pval)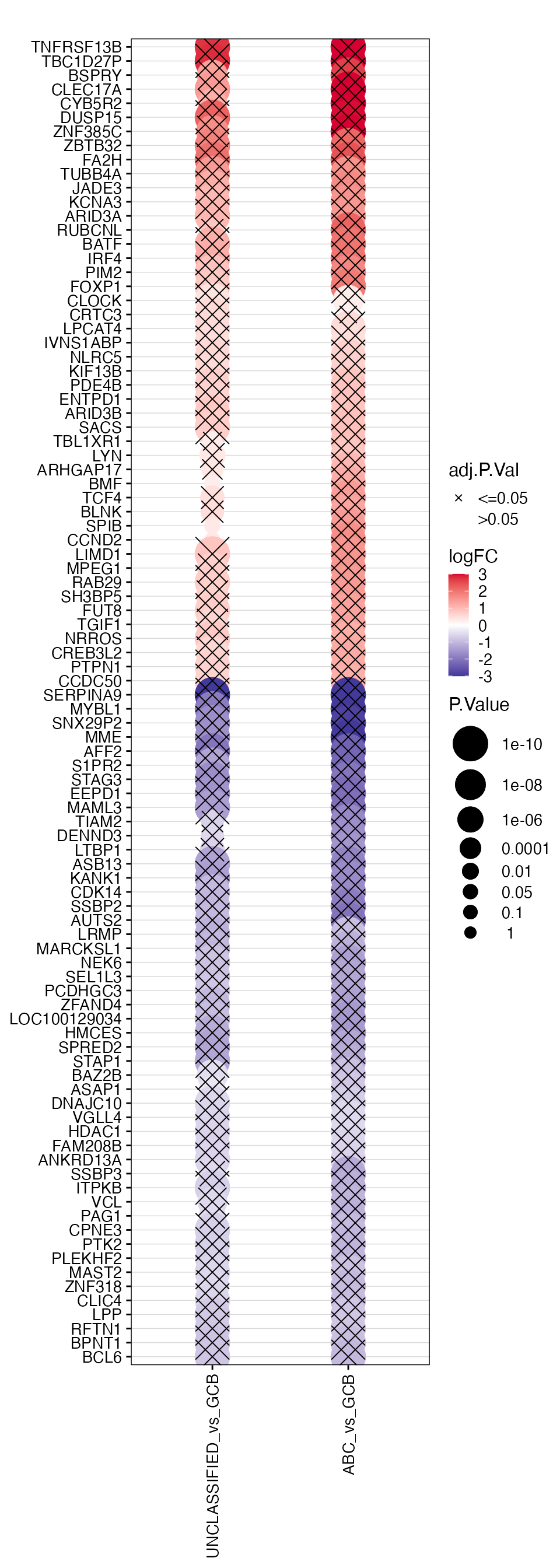
### logFC_heatmap
wrapper_logFC_heatmap(topTable_significant_genes, gene_var = "HGNC_Symbol", lfc_prefix = "logFC",
adjp_prefix = "adj.P.Val", cluster_rows = cluster_rows, row_names_gp = grid::gpar(fontsize = 7))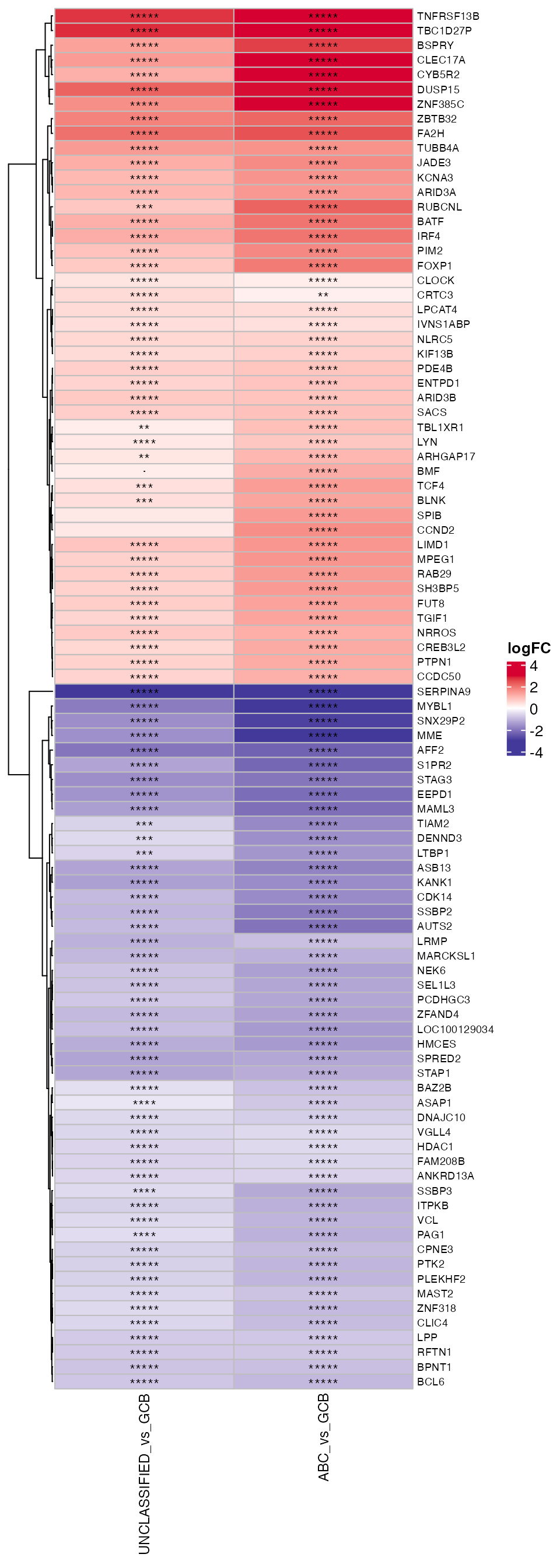
### z-score heatmap
expr_de <- v$E[significant_genes, , drop = FALSE]
wrapper_gene_expression_heatmap(expr_de, cluster_rows = cluster_rows, column_split = pdata[,
group_var], row_names_gp = grid::gpar(fontsize = 7), trim_prop = 0.05)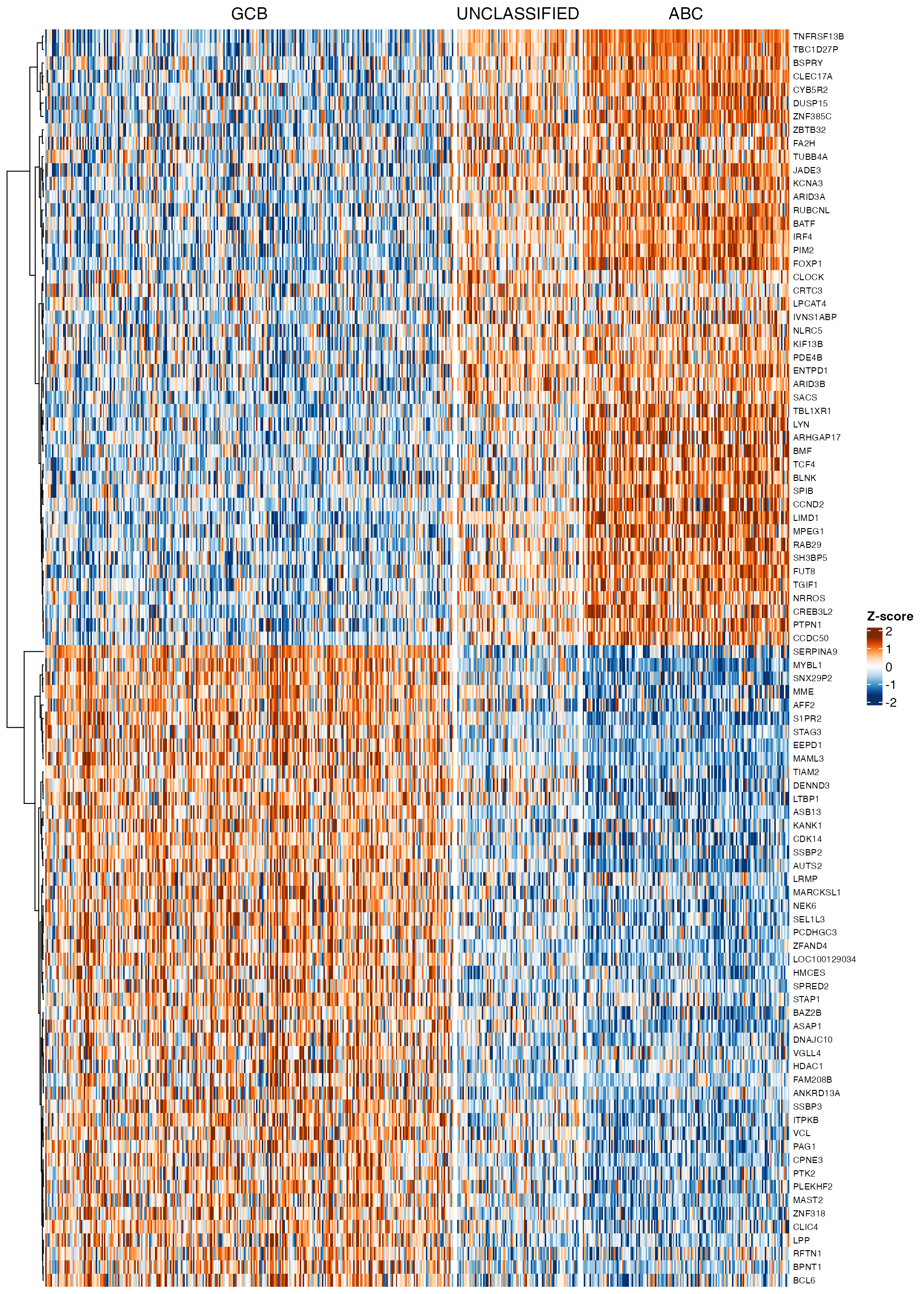
### Mean z-score heatmap
data_adjp <- wrapper_extract_from_topTable(topTable_significant_genes, extract_prefix = "adj.P.Val")
data_adjp <- data_adjp[, names(contrast_mapping), drop = FALSE]
colnames(data_adjp) <- contrast_mapping
wrapper_summarized_expression_heatmap(expr_de, group = pdata[, group_var], adjp = data_adjp,
cluster_rows = cluster_rows, row_names_gp = grid::gpar(fontsize = 7), trim_values = 1)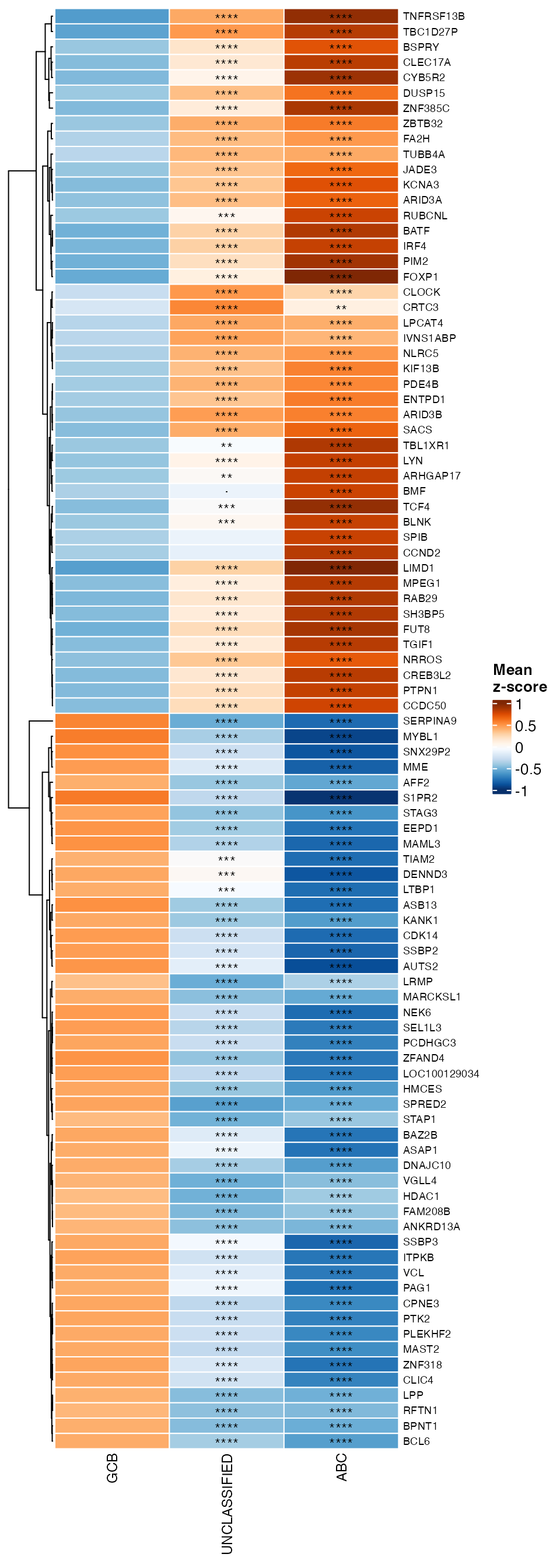
Gene set enrichment analysis (GSEA)
gene_mapping <- NULL
gene_var <- "HGNC_Symbol"
genesets <- geneset_list_hallmark
genesets_extra_info <- geneset_extra_hallmark
geneset_vars <- "GenesetID"
############################################################################# Run
############################################################################# GSEA
set.seed(1234)
topTable_gsea <- wrapper_gsea(topTable, genesets = genesets, genesets_extra_info = genesets_extra_info,
gene_mapping = gene_mapping, gene_var = gene_var, topn_genes = Inf)
rownames(topTable_gsea) <- topTable_gsea$GenesetID
head(topTable_gsea)## GenesetID Category Size_UNCLASSIFIED_vs_GCB
## ADIPOGENESIS ADIPOGENESIS HALLMARK 192
## ALLOGRAFT REJECTION ALLOGRAFT REJECTION HALLMARK 178
## ANDROGEN RESPONSE ANDROGEN RESPONSE HALLMARK 95
## ANGIOGENESIS ANGIOGENESIS HALLMARK 32
## APICAL JUNCTION APICAL JUNCTION HALLMARK 183
## APICAL SURFACE APICAL SURFACE HALLMARK 40
## Leading.Genes_UNCLASSIFIED_vs_GCB
## ADIPOGENESIS BCL6, PFKL, CPT2, CDKN2C, CCNG2, SOD1, COQ5, UBQLN1, CS, ALDH2, PHYH, DBT, NDUFA5, ATL2, MRPL15, ACO2, C3, ACADM, GPAT4, AK2, DNAJC15, COX7B, MAP4K3, MDH2, PGM1, COX6A1, LPL, SULT1A1, NMT1, TANK, APLP2, DLD, TKT, ATP5PO, DRAM2, PHLDB1, SLC25A1, FABP4, OMD, HSPB8, ADIPOR2
## ALLOGRAFT REJECTION IRF4, IL16, IL12A, IL2RB, LYN, IFNAR2, CCL22, IL10, BCL3, IL12B, IL1B, GZMB, CD47, GALNT1, IL6, GBP2, TAP1, CXCL13, IFNG, IL7, PRKCG, PRF1, ZAP70, TAP2, FGR, AKT1, PRKCB, NCR1, APBB1, IL2RA, CCR1, HLA-G, GPR65, B2M, CCND2, CD80, CXCR3, CCR5, TLR2, BCL10, PSMB10, STAT1, SRGN, GLMN, CD7, CD96, SOCS5, PTPN6, CCL2, FLNA, KLRD1, JAK2, CXCL9, STAB1, CD1D, IL27RA, CD8B, C2, ETS1, ITGAL, HLA-A, MRPL3, NLRP3, DYRK3, CD8A
## ANDROGEN RESPONSE ABCC4, PTK2B, TPD52, ALDH1A3, DBI, PMEPA1, UBE2J1, CAMKK2, SMS, ELOVL5, STK39, HERC3, RAB4A, ANKH, ITGAV
## ANGIOGENESIS PTK2, OLR1, POSTN, MSX1, ITGAV, LUM, FSTL1, LPL, TIMP1, SPP1, JAG2, COL5A2, COL3A1, KCNJ8, FGFR1
## APICAL JUNCTION PTK2, VCL, EPB41L2, CD86, IRS1, ACTN2, MAP4K2, AMH, NF1, SLIT2, EVL, EXOC4, LIMA1, TSC1, MAPK14, SYK, LAYN, FBN1, MMP9, CAP1, SHROOM2, FSCN1, COL16A1, MSN, THY1, SKAP2, PTPRC, AMIGO2, RHOF, TNFRSF11B, CDH11, JUP, PARVA, CDH6, RSU1, ACTG1, STX4, ATP1A3, WASL, ITGB1, TRAF1, RASA1, EGFR, DSC3
## APICAL SURFACE IL2RB, LYN, SLC2A4, CD160, ADAM10, B4GALT1, MDGA1, NCOA6, ATP6V0A4, ATP8B1, APP
## Median.t_UNCLASSIFIED_vs_GCB Direction_UNCLASSIFIED_vs_GCB
## ADIPOGENESIS -0.28 Down
## ALLOGRAFT REJECTION 0.64 Up
## ANDROGEN RESPONSE 0.13 Down
## ANGIOGENESIS -0.75 Down
## APICAL JUNCTION 0.02 Down
## APICAL SURFACE 0.00 Up
## ES_UNCLASSIFIED_vs_GCB NES_UNCLASSIFIED_vs_GCB
## ADIPOGENESIS -0.20 -0.95
## ALLOGRAFT REJECTION 0.36 1.70
## ANDROGEN RESPONSE -0.22 -0.89
## ANGIOGENESIS -0.31 -1.02
## APICAL JUNCTION -0.24 -1.09
## APICAL SURFACE 0.26 0.95
## P.Value_UNCLASSIFIED_vs_GCB adj.P.Val_UNCLASSIFIED_vs_GCB
## ADIPOGENESIS 0.6039783002 0.7107286177
## ALLOGRAFT REJECTION 0.0001181803 0.0006565574
## ANDROGEN RESPONSE 0.6563706564 0.7313368056
## ANGIOGENESIS 0.4291262136 0.6310679612
## APICAL JUNCTION 0.2477558348 0.4129263914
## APICAL SURFACE 0.5679012346 0.7107286177
## sign.P.Val_UNCLASSIFIED_vs_GCB Size_ABC_vs_GCB
## ADIPOGENESIS -0.2189787 192
## ALLOGRAFT REJECTION 3.9274548 178
## ANDROGEN RESPONSE -0.1828508 95
## ANGIOGENESIS -0.3674150 32
## APICAL JUNCTION -0.6059761 183
## APICAL SURFACE 0.2457272 40
## Leading.Genes_ABC_vs_GCB
## ADIPOGENESIS ATP1B3, IDH3A, DLAT, MCCC1, RREB1, PDCD4, UCP2, ESYT1, ACAA2, GPD2, SLC1A5, ACADM, JAGN1, NDUFAB1, IMMT, QDPR, NMT1, SLC19A1, BAZ2A, GRPEL1, UQCRC1, SLC25A10, COQ3, NDUFS3, SDHC, HADH, NKIRAS1, ECHS1, BCL2L13, NABP1, ACLY, MRPL15, YWHAG, CD36, AGPAT3, ECH1, CYC1, PEX14, GHITM, MDH2, SAMM50, DECR1, UQCRQ, TALDO1, MTCH2, ACADS, NDUFB7, AIFM1, UQCR11, PTCD3, CHCHD10
## ALLOGRAFT REJECTION IRF4, LYN, CCND2, IL16, IL12A, MRPL3, CD47, ABCE1, PRKCB, IL10, IL7, GZMB, NPM1, ETS1, NME1, BCL3, SIT1, IL2RA, CSK, RPS9, PSMB10, GLMN, ABI1
## ANDROGEN RESPONSE PTK2B, PMEPA1, HERC3, ITGAV, ABCC4, SGK1, PTPN21, GUCY1A1, CAMKK2, ACTN1, ALDH1A3, CCND3, TPD52, STK39, CDC14B, CCND1, PLPP1, FADS1, ELOVL5, MAK, MERTK, HPGD, TSC22D1, BMPR1B, ZBTB10, MAF, ABHD2, PIAS1, MAP7, UBE2J1, HSD17B14, KRT19
## ANGIOGENESIS PTK2, ITGAV, POSTN, LUM, COL3A1, COL5A2, FSTL1, OLR1, FGFR1, VCAN, JAG1, JAG2, MSX1
## APICAL JUNCTION PTK2, VCL, EPB41L2, MMP2, CDH11, FBN1, COL16A1, EVL, PARVA, NF1, NLGN2, BMP1, EGFR, CERCAM, ACTN2, ACTN1, MYH9, JUP, SHC1, VCAN, TJP1, NECTIN3, FSCN1, PLCG1, WASL, LIMA1, THY1, CTNND1, THBS3, CDH6, LAMB3, IRS1, ADAM23, SHROOM2, MMP9, MYL9, MAPK14, MAPK13, CD276, ADAMTS5, CD86, SLIT2, TGFBI, GNAI1, GAMT, SIRPA, MSN, FLNC, BAIAP2, TRAF1, RSU1, CD99, JAM3, PCDH1, RHOF, SPEG, NEXN, STX4, ATP1A3, COL9A1, NEGR1, ICAM1, ALOX15B, AMH, ITGB1, TSC1, RRAS, CDH8, PTPRC, MDK, ZYX, CDH1, NLGN3, CDH3, NECTIN4, LAMC2, NFASC, SRC, CRAT, AKT3, DSC3, MAP4K2, MYH10, FYB1, EXOC4, LAYN, CTNNA1, TNFRSF11B, PKD1, LDLRAP1
## APICAL SURFACE EFNA5, CRYBG1, AKAP7, THY1, PKHD1, SHROOM2, DCBLD2, GAS1, GHRL, PLAUR, GSTM3, HSPB1, AFAP1L2, LYPD3, NTNG1, CROCC, ATP8B1, SRPX, MDGA1, GATA3, SCUBE1
## Median.t_ABC_vs_GCB Direction_ABC_vs_GCB ES_ABC_vs_GCB
## ADIPOGENESIS -0.33 Up 0.17
## ALLOGRAFT REJECTION -0.22 Up 0.28
## ANDROGEN RESPONSE -1.30 Down -0.28
## ANGIOGENESIS -3.49 Down -0.56
## APICAL JUNCTION -2.30 Down -0.42
## APICAL SURFACE -2.96 Down -0.41
## NES_ABC_vs_GCB P.Value_ABC_vs_GCB adj.P.Val_ABC_vs_GCB
## ADIPOGENESIS 0.81 9.153094e-01 9.153094e-01
## ALLOGRAFT REJECTION 1.30 4.038772e-02 7.479208e-02
## ANDROGEN RESPONSE -1.24 1.083123e-01 1.692380e-01
## ANGIOGENESIS -2.00 3.674254e-04 1.224751e-03
## APICAL JUNCTION -2.10 3.324609e-09 1.385254e-08
## APICAL SURFACE -1.56 1.686440e-02 3.372879e-02
## sign.P.Val_ABC_vs_GCB
## ADIPOGENESIS 0.03843206
## ALLOGRAFT REJECTION 1.39375064
## ANDROGEN RESPONSE -0.96532205
## ANGIOGENESIS -3.43483079
## APICAL JUNCTION -8.47825941
## APICAL SURFACE -1.77302921
# ------------------------------------------------------------ Generate tables
# with significant results
# ------------------------------------------------------------
significant_gsea_list <- lapply(1:length(contrasts), function(i) {
# i = 1
contrast <- contrasts[i]
cat(paste0("\n\n### GSEA results for ", gsub("_", " ", contrast), "\n\n"))
# ------------------------------------------- up
# -------------------------------------------
direction <- "up"
topTable_up <- wrapper_dispaly_significant_gsea(topTable_gsea, contrast = contrast,
direction = direction, topn = 20, pval = pval, geneset_vars = geneset_vars,
direction_prefix = "Direction", pval_prefix = "P.Value", adjp_prefix = "adj.P.Val")
print(topTable_up)
### Plot
if (nrow(topTable_up) > 0) {
genesets2plot <- genesets[bresults(topTable_up)$GenesetID]
names(genesets2plot) <- bresults(topTable_up)$GenesetID
title <- bcaption(topTable_up)
gsea_results <- bresults(topTable_up)
ggp <- wrapper_gsea_plot(topTable, genesets = genesets2plot, contrast = contrast,
title = title, gene_var = gene_var, gsea_results = gsea_results, geneset_var = "GenesetID")
print(ggp)
}
# ------------------------------------------- down
# -------------------------------------------
direction <- "down"
topTable_down <- wrapper_dispaly_significant_gsea(topTable_gsea, contrast = contrast,
direction = direction, topn = 20, pval = pval, geneset_vars = geneset_vars,
direction_prefix = "Direction", pval_prefix = "P.Value", adjp_prefix = "adj.P.Val")
print(topTable_down)
### Plot
if (nrow(topTable_down) > 0) {
genesets2plot <- genesets[bresults(topTable_down)$GenesetID]
names(genesets2plot) <- bresults(topTable_down)$GenesetID
title <- bcaption(topTable_down)
gsea_results <- bresults(topTable_down)
ggp <- wrapper_gsea_plot(topTable, genesets = genesets2plot, contrast = contrast,
title = title, gene_var = gene_var, gsea_results = gsea_results, geneset_var = "GenesetID")
print(ggp)
}
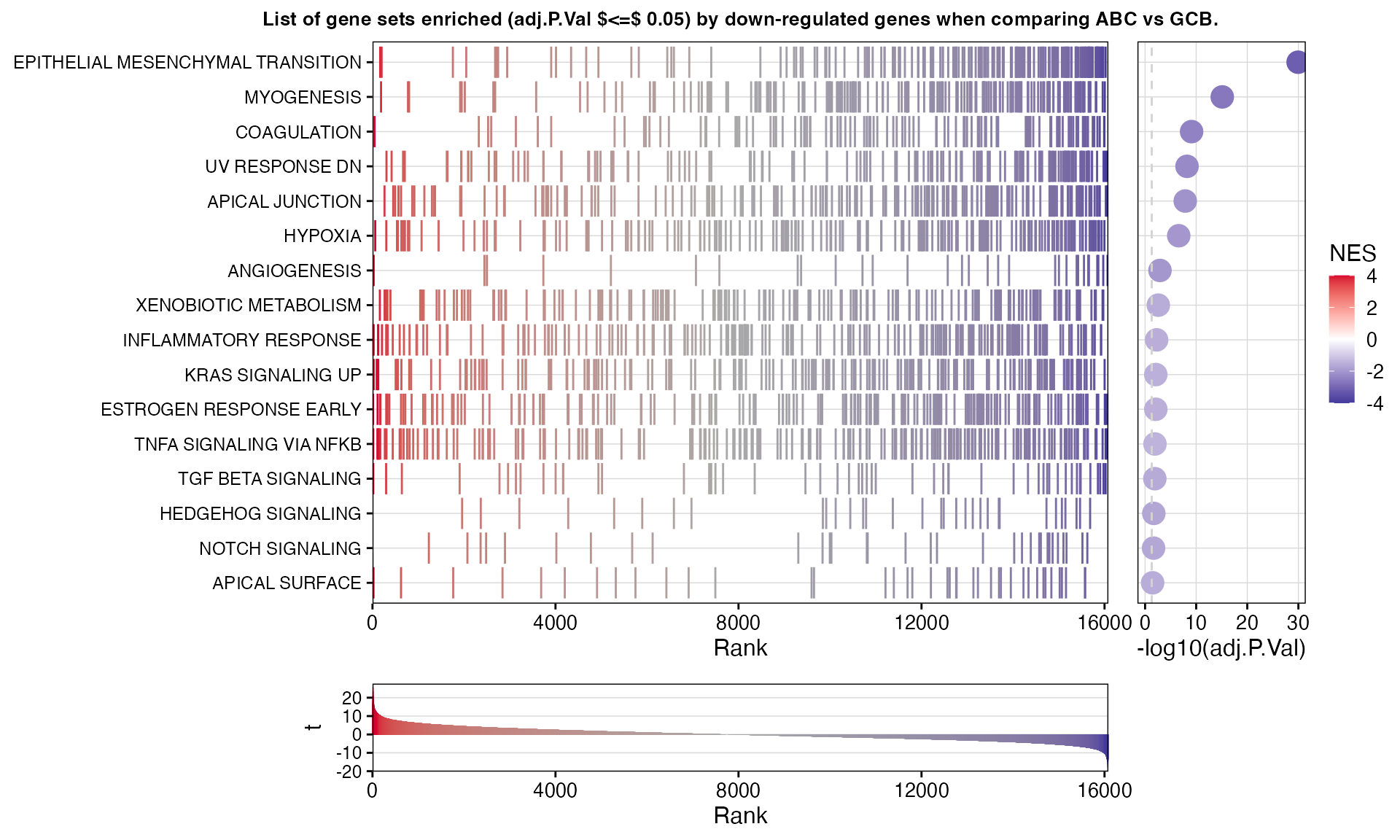
significant_gsea <- c(bresults(topTable_up)$GenesetID, bresults(topTable_down)$GenesetID)
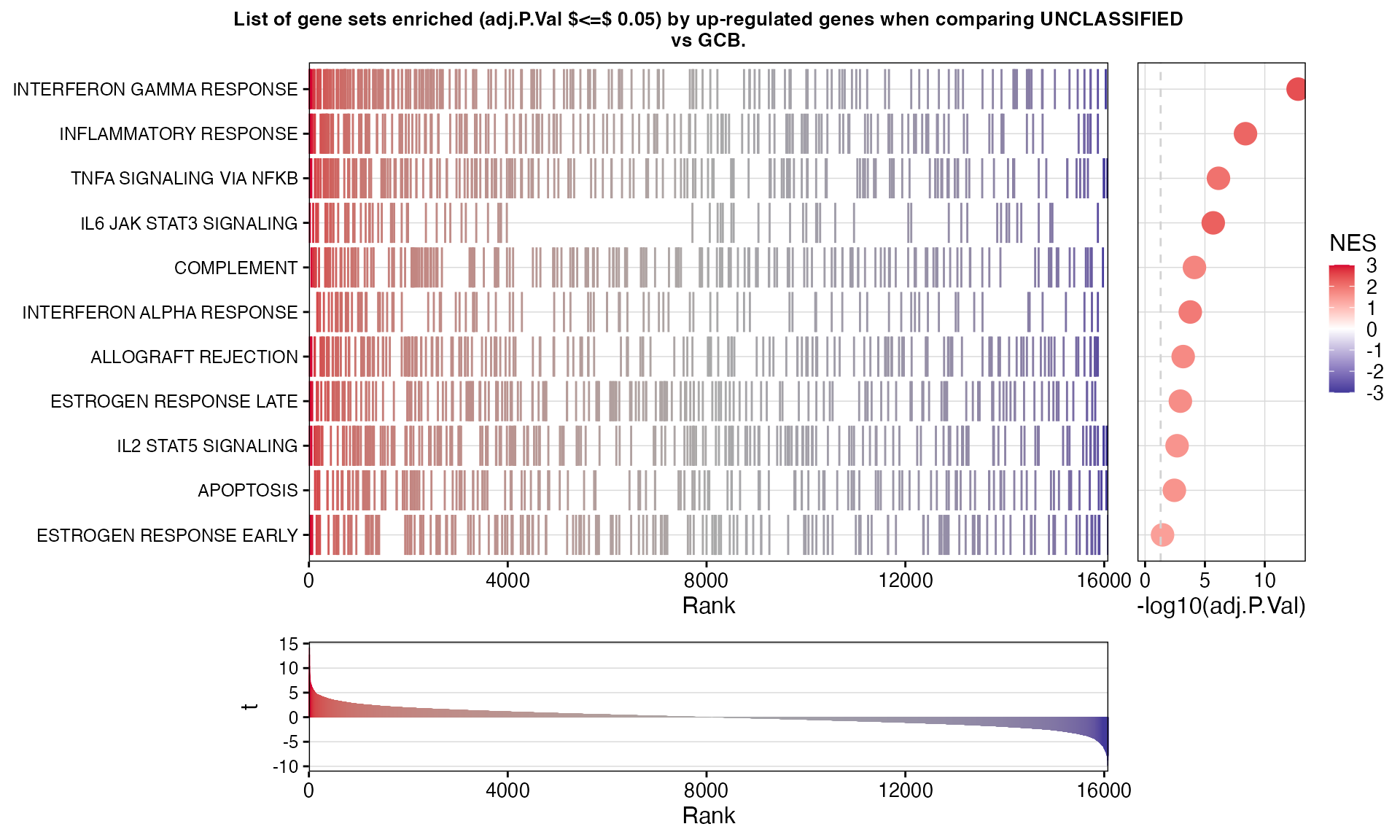
})GSEA results for UNCLASSIFIED vs GCB
| GenesetID | Direction | Size | Leading.Genes | NES | P.Value | adj.P.Val |
|---|---|---|---|---|---|---|
| INTERFERON GAMMA RESPONSE | Up | 195 | IRF4, PDE4B, PTPN1, NLRC5, IL10RA, PTPN2, RNF213, IL2RB, SP110, ST3GAL5, TRIM21, CASP7, PNP, SAMD9L, PML, IFNAR2, NMI, IL15RA, MT2A, ITGB7, CASP4, SECTM1, SOCS3, IL6, STAT3, BATF2, TAP1, NAMPT, IRF2, CASP8, BPGM, PTGS2, IDO1, MYD88, FPR1, IL7, IRF1, SLAMF7, CD69, PIM1, FGL2, CASP1, IFITM2, CXCL10, CD274, CFB, CXCL11, SLC25A28, FCGR1A, SOD2, IL18BP, HLA-G, VAMP5, ZNFX1, METTL7B, B2M, UBE2L6, BANK1, DDX58, RTP4, PSMB8, PFKP, DDX60, TRIM25, NOD1, PSME2, NFKBIA, PSMB10, STAT1, MTHFD2, EIF4E3, PTPN6, CCL2, TNFSF10, JAK2, TNFAIP2, CXCL9, PLA2G4A, TRAFD1, TNFAIP3, CDKN1A, APOL6, PSME1, ZBP1, RNF31, GBP4, IRF5 | 2.46 | <1e-14 *** | <1e-12 *** |
| INFLAMMATORY RESPONSE | Up | 188 | KCNA3, PDE4B, IL10RA, CCR7, CHST2, IL2RB, LYN, PTGER4, CCL22, IL10, GPR183, NMI, IL15RA, KCNJ2, EBI3, IL12B, FFAR2, MEFV, IL1B, SEMA4D, PROK2, ADGRE1, PTPRE, IL6, NAMPT, SELENOS, FPR1, SELL, IRF1, CSF3R, P2RY2, IL1A, CD69, CCL24, CXCL10, NOD2, MET, PTGER2, CXCL11, RASGRP1, KIF1B, TNFRSF1B, MARCO, AQP9, RTP4, CXCL6, PIK3R5, ADM, SLC31A2, TLR2, NFKBIA, LDLR, OSM, GNA15, CCL2, TNFSF10, CD55, CXCL9, MYC, CDKN1A, STAB1, HAS2, CD48, CXCR6 | 2.18 | <1e-09 *** | <1e-08 *** |
| TNFA SIGNALING VIA NFKB | Up | 196 | PDE4B, TGIF1, CFLAR, CXCL2, KLF2, CD44, LITAF, PTGER4, GPR183, BCL3, ID2, IL15RA, DUSP5, CXCL3, IL12B, IL1B, TNFAIP8, ZFP36, CXCL1, SOCS3, PTPRE, IL6, TRIB1, IER5, TAP1, NAMPT, PTGS2, IRF1, IL1A, CD69, SERPINB8, MARCKS, CSF2, EGR3, CXCL10, CCNL1, DRAM1, NINJ1, NR4A2, CXCL11, SOD2, EHD1, DDX58, IER3, CD80, MCL1, CXCL6, PHLDA1, NR4A3, SLC2A3, NFKBIE, CEBPB, MAP2K3, TLR2, NFKBIA, B4GALT1, LDLR, JUN, AREG, SAT1, CCL2, PTX3, TNFAIP2, TNFAIP3, MYC, CDKN1A, PANX1, BHLHE40, DENND5A, FUT4, GCH1, MXD1, ATF3, ZC3H12A, PER1, CCL4, IL7R, VEGFA, CCRL2, KLF6, LIF, G0S2, RCAN1, BMP2, STAT5A, PPP1R15A, IFIT2, KLF4 | 2.02 | <1e-07 *** | <1e-06 *** |
| IL6 JAK STAT3 SIGNALING | Up | 78 | PTPN1, PTPN2, IL10RB, CD44, IL15RA, EBI3, CXCL3, IL1B, CXCL1, SOCS3, IL6, STAT3, CXCL13, MYD88, IL7, BAK1, IRF1, IL17RB, CSF3R, PIM1, CSF2, CXCL10, CXCL11, TNFRSF1B, IL2RA, CCR1, IL17RA, HMOX1, ITGA4, PIK3R5, LTBR, TLR2, JUN, STAT1 | 2.25 | <1e-06 *** | <1e-05 *** |
| COMPLEMENT | Up | 190 | TMPRSS6, CASP5, DGKG, LYN, DOCK10, MSRB1, CASP7, GPD2, DUSP5, CASP4, GZMB, APOBEC3G, CASP10, CXCL1, IL6, S100A12, IRF2, DPP4, S100A9, IRF1, KLK1, PIM1, C4BPB, USP8, CASP1, CFB, RASGRP1, EHD1, C1QA, CP, F5, PIK3R5, ANG, CEBPB, C1QC, CTSC, ME1, ANXA5, SCG3, SERPINC1, LGALS3, KCNIP2, CLU, SERPINA1, JAK2, DGKH, CD55, CTSB, PLA2G4A, TNFAIP3, SPOCK2, CBLB, FCER1G, CTSV, C2 | 1.77 | <1e-05 *** | <1e-04 *** |
| INTERFERON ALPHA RESPONSE | Up | 96 | SP110, TRIM21, SAMD9L, NMI, CD47, MOV10, GBP2, BATF2, TAP1, SAMD9, IRF2, CASP8, IL7, SELL, IRF1, PARP9, CASP1, IFITM2, CXCL10, UBA7, CXCL11, SLC25A28, B2M, TENT5A, UBE2L6, RTP4, PSMB8, DDX60, TRIM25, PSME2, TRAFD1, PSME1, RNF31, HLA-C, GBP4, HELZ2, IFITM3, IRF9, ISG20, TRIM5, PSMA3, CCRL2 | 1.91 | <1e-04 *** | 0.0002 *** |
| ALLOGRAFT REJECTION | Up | 178 | IRF4, IL16, IL12A, IL2RB, LYN, IFNAR2, CCL22, IL10, BCL3, IL12B, IL1B, GZMB, CD47, GALNT1, IL6, GBP2, TAP1, CXCL13, IFNG, IL7, PRKCG, PRF1, ZAP70, TAP2, FGR, AKT1, PRKCB, NCR1, APBB1, IL2RA, CCR1, HLA-G, GPR65, B2M, CCND2, CD80, CXCR3, CCR5, TLR2, BCL10, PSMB10, STAT1, SRGN, GLMN, CD7, CD96, SOCS5, PTPN6, CCL2, FLNA, KLRD1, JAK2, CXCL9, STAB1, CD1D, IL27RA, CD8B, C2, ETS1, ITGAL, HLA-A, MRPL3, NLRP3, DYRK3, CD8A | 1.70 | 0.0001 *** | 0.0007 *** |
| ESTROGEN RESPONSE LATE | Up | 182 | BATF, AFF1, ITPK1, SLC9A3R1, CD44, TST, ID2, LLGL2, ZFP36, MYB, NMU, SLC27A2, GALE, XBP1, CHPT1, ALDH3A2, PDCD4, FOXC1, S100A9, IL17RB, OPN3, EGR3, HMGCS2, ARL3, CAV1, BLVRB, FABP5, SLC26A2, PRLR, DNAJC1, ALDH3B1, LARGE1, AREG, PKP3, TJP3, TFF3, PTPN6, AGR2, SERPINA1, CYP4F11, JAK2, ASS1, KRT19, IMPA2, RAB31, CELSR2, PTGES, WFS1, CDH1, SORD, MYOF, UGDH, RPS6KA2, RET, LTF, ISG20, ANXA9, MDK, GPER1, SLC2A8, SULT2B1, KLF4, CA2, JAK1, KLK11, RAPGEFL1, CA12, SLC7A5, BTG3, HR | 1.65 | 0.0002 *** | 0.0011 ** |
| IL2 STAT5 SIGNALING | Up | 195 | BATF, IRF4, IL10RA, IL2RB, NFKBIZ, CAPN3, CD44, IGF2R, ENO3, PNP, AHNAK, IL10, UCK2, XBP1, HIPK2, MUC1, SELL, HK2, PIM1, FGL2, CSF2, FURIN, FLT3LG, CKAP4, CXCL10, PTGER2, RORA, ADAM19, TNFRSF1B, IL2RA, COCH, PLAGL1, GPR65, LCLAT1, CCND2, SLC1A5, PHLDA1, BMPR2, BATF3, SLC2A3, MAPKAPK2, TTC39B, TGM2, AMACR, SERPINC1, TNFSF10, LRRC8C, CYFIP1, MYC, SLC29A2, ETV4, CD48, BHLHE40, GBP4, DENND5A, TNFRSF21, CCR4, CDC42SE2, IFITM3, MXD1, CST7, SOCS2 | 1.55 | 0.0005 *** | 0.0021 ** |
| APOPTOSIS | Up | 156 | CFLAR, PAK1, CD44, IGF2R, CASP7, RARA, BCL2L10, IL1B, CASP4, IL6, TAP1, BID, CASP8, PDCD4, IRF1, IL1A, PRF1, CD69, EGR3, DNAJC3, CASP1, RETSAT, PLCB2, CAV1, AIFM3, SOD2, BMF, HMOX1, IER3, CCND2, MCL1, TGFBR3, CREBBP, GPX1, CDKN1B, BCL10, JUN, SAT1, GNA15, LGALS3, KRT18, SLC20A1, CLU, TNFSF10, ADD1, CDKN1A, CTH, CDC25B, GCH1, IFITM3, NEFH, CD14, ATF3, ISG20, LEF1, HGF, APP, CYLD, RNASEL, FEZ1, BMP2 | 1.56 | 0.0009 *** | 0.0036 ** |
| ESTROGEN RESPONSE EARLY | Up | 185 | FCMR, AFF1, SH3BP5, ITPK1, UNC119, SLC9A3R1, CD44, RARA, GAB2, MYB, SLC27A2, SLC19A2, ABAT, XBP1, CHPT1, MUC1, FOXC1, IL17RB, P2RY2, OPN3, KRT8, EGR3, ARL3, RASGRP1, ELF3, BLVRB, RHOD, RRP12, SLC26A2, SYNGR1, RHOBTB3, B4GALT1, ALDH3B1, AREG, WWC1, TJP3, TGM2, TFF3, KRT18, FASN, JAK2, MLPH, MYC, ADCY1, CLDN7, KRT19, BHLHE40, RAB31, MPPED2, CELSR2, PTGES, WFS1, TTC39A | 1.40 | 0.0104 * | 0.0348 * |
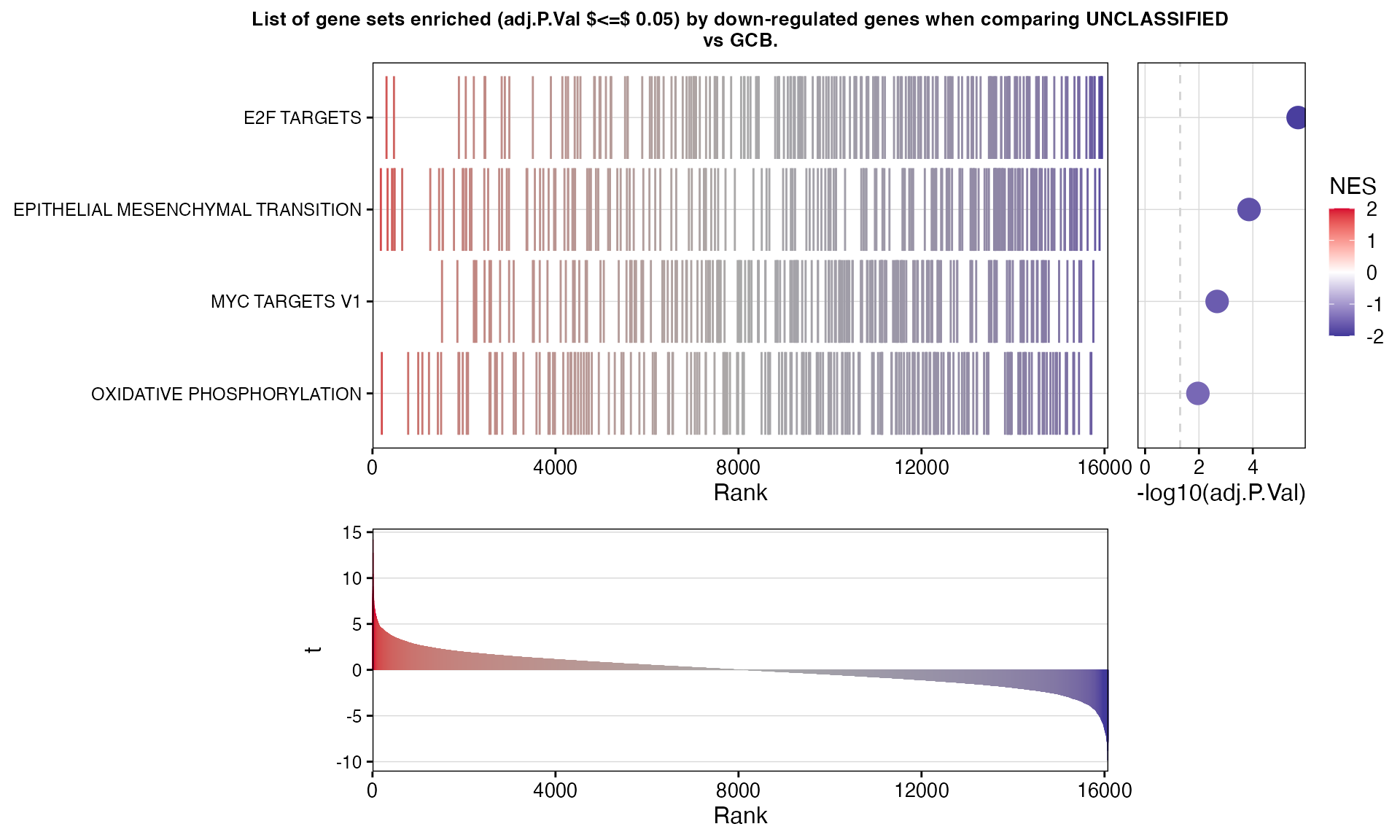
| GenesetID | Direction | Size | Leading.Genes | NES | P.Value | adj.P.Val |
|---|---|---|---|---|---|---|
| E2F TARGETS | Down | 198 | WEE1, DCK, DNMT1, PSIP1, CDKN2C, RFC2, RBBP7, PTTG1, RFC1, PRPS1, NAP1L1, ING3, HUS1, NCAPD2, DEPDC1, CBX5, BRMS1L, TIMELESS, RPA3, RACGAP1, POLA2, LBR, NUP107, EZH2, NUP205, HMGA1, SLBP, AK2, RAN, UBR7, PRIM2, CDK1, CDKN3, SMC1A, TK1, UNG, RFC3, XPO1, MSH2, RNASEH2A, CDKN2A, PMS2, DEK, STMN1, PRKDC, ASF1A, PHF5A, MCM7, UBE2T, RPA2, CDCA3, RAD21, BIRC5, SMC3, CIT, TP53, DONSON, CENPM, CCNB2, GINS3, BARD1, HMMR, RAD51C, ORC2, HMGB3, MRE11, MLH1, POLD2, SNRPB, NAA38, CKS2, SMC4, KIF4A, ATAD2, PPP1R8, DSCC1, LUC7L3, CHEK2, PAN2, CSE1L, MCM6, MCM4, PA2G4, CTCF | -1.94 | <1e-06 *** | <1e-05 *** |
| EPITHELIAL MESENCHYMAL TRANSITION | Down | 194 | TIMP3, BASP1, LRRC15, POSTN, FBLN2, FN1, COMP, BGN, WIPF1, PMEPA1, ADAM12, CTHRC1, SLIT2, FAP, DPYSL3, TFPI2, THBS2, MXRA5, ECM2, FZD8, MSX1, LAMA2, COL11A1, GLIPR1, LOXL2, SPARC, FBN1, DCN, PLOD2, ITGAV, NNMT, SFRP4, CRLF1, HTRA1, ELN, FBLN5, MATN3, LUM, COL16A1, FSTL1, MFAP5, THY1, COL6A3, GADD45B, MEST, PFN2, SPOCK1, COL5A1, TIMP1, FSTL3, TPM1, TNFRSF11B, SERPINE2, COL12A1, SGCD, CDH11, MCM7, LOXL1, CDH6, COL1A2, PMP22, EDIL3, SPP1, COL1A1, ITGB1, SNAI2, FERMT2, GEM, PCOLCE2, CALD1, COL8A2, MGP, LOX, PCOLCE, ABI3BP, FBLN1, IGFBP2, IGFBP3, COL5A2, CALU, ECM1, MMP2, COL3A1, DST, SGCB | -1.72 | <1e-04 *** | 0.0001 *** |
| MYC TARGETS V1 | Down | 193 | HDDC2, UBE2E1, NAP1L1, CDK2, CSTF2, NCBP1, RRM1, SSB, SSBP1, EIF4H, PABPC4, RPL6, YWHAQ, DDX18, PHB2, UBA2, RAN, G3BP1, HPRT1, PSMA2, LDHA, CCT4, STARD7, VBP1, XPO1, ACP1, DEK, CBX3, RACK1, RPS6, CLNS1A, MCM7, COPS5, TARDBP, CCT2, PSMD14, RPL14, SRSF3, PGK1, POLE3, RPL34, MRPS18B, SLC25A3, ORC2, C1QBP, CCT7, EIF3B, LSM7, POLD2, EIF1AX, YWHAE, AIMP2, PSMA7, PABPC1, HNRNPR, XPOT, PRDX3, HNRNPU, NHP2, IFRD1, RPS3, MCM6, MCM4, PA2G4, SNRPD2, RPL22, ODC1, EEF1B2, RAD23B, NCBP2, CDC20, HSPD1, CYC1, TFDP1, EXOSC7, CCT3, IMPDH2, CDK4, KPNB1, PSMD7, MRPL9, PSMB3, HNRNPA2B1, PSMA6, PPM1G, KPNA2, PSMB2, PSMD3, RPS5, HNRNPA3, HNRNPA1, PRPS2, XRCC6, TUFM, RANBP1, PSMD1, CAD, SNRPB2, PTGES3 | -1.61 | 0.0005 *** | 0.0021 ** |
| OXIDATIVE PHOSPHORYLATION | Down | 200 | MDH1, NDUFB2, COX15, ATP6V1G1, CS, ABCB7, UQCRB, ATP5MG, PHYH, OAT, NDUFA5, CYB5R3, COX11, MRPL15, ACO2, ACADM, ATP5PB, COX7A2L, PHB2, COX4I1, NDUFA8, COX7B, FXN, TOMM22, LDHA, NDUFB3, MDH2, SLC25A5, NDUFA2, COX6A1, COX17, NDUFB1, SLC25A4, UQCRH, MRPL35, ATP6V1H, NDUFA4, NDUFS7, MTX2, PDHA1, ATP5MF, DLD, ATP5PF, ATP5PO, COX7C, ATP5MC1, NDUFB6, NDUFA1, ATP5MC2, TIMM13, FDX1, ATP6V0E1, MRPL11, ATP6V1F, COX8A, SLC25A3, COX6C, SLC25A20, UQCRQ, ATP5MC3, BDH2, BCKDHA, HSD17B10, ATP6V1C1, ATP5PD, NQO2, ATP5F1C, MPC1, NDUFB5, ACAT1, POLR2F, PRDX3, NDUFC1, ETFB, NDUFA7, NNT, NDUFV1, ATP5F1B, MTRF1, IDH3G, TIMM17A, NDUFS1, PMPCA, ATP6V1D | -1.48 | 0.0031 ** | 0.0109 * |
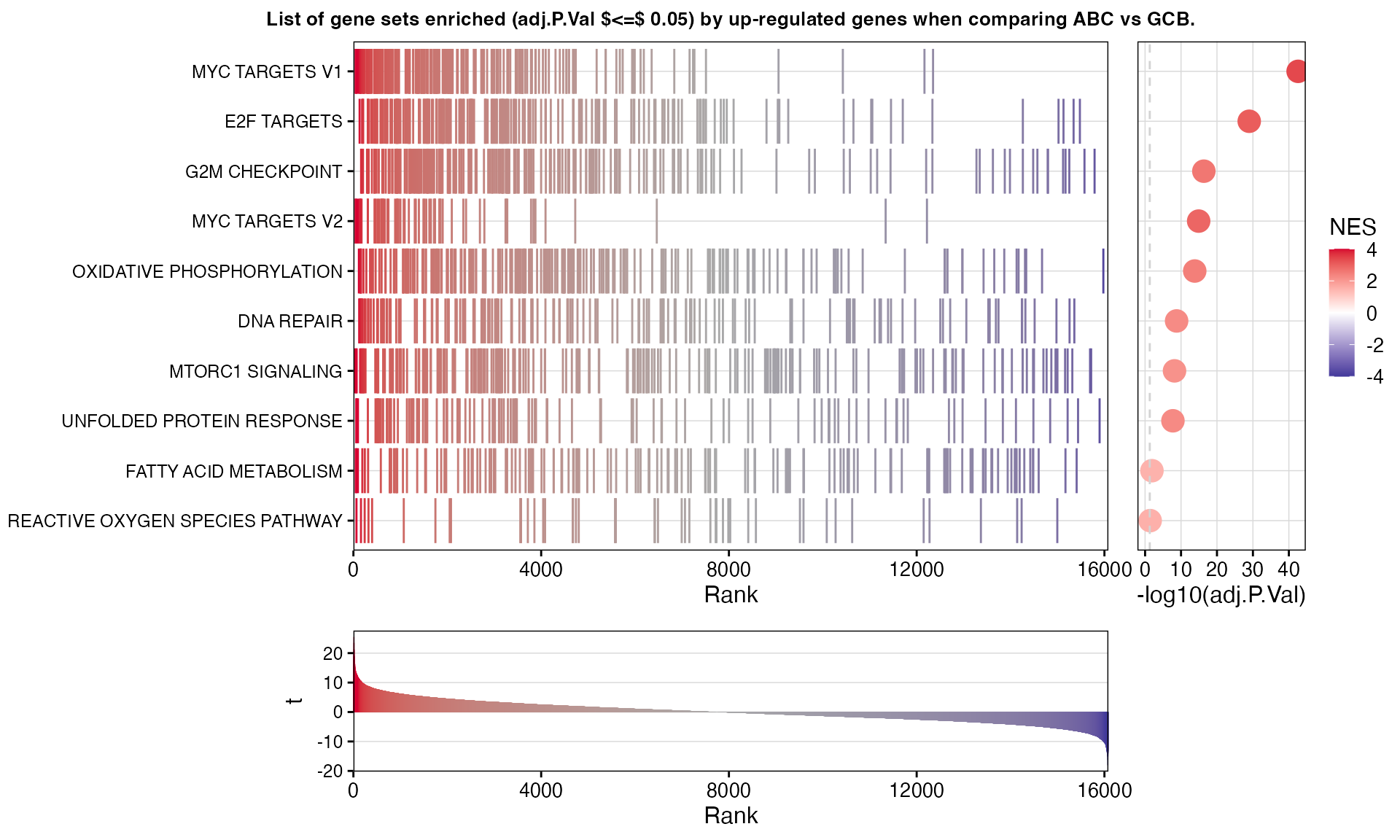
GSEA results for ABC vs GCB
| GenesetID | Direction | Size | Leading.Genes | NES | P.Value | adj.P.Val |
|---|---|---|---|---|---|---|
| MYC TARGETS V1 | Up | 193 | GNL3, RRP9, APEX1, RFC4, SRM, TOMM70, BUB3, MCM2, ABCE1, CNBP, TXNL4A, SNRPA, FBL, MYC, NOLC1, TRA2B, SET, GOT2, TYMS, PWP1, RSL1D1, MCM5, RUVBL2, EXOSC7, RNPS1, RPS5, ILF2, USP1, NCBP2, TRIM28, SNRPD1, NPM1, RPL18, DUT, SF3B3, IMPDH2, HSP90AB1, RPL14, SMARCC1, EIF4G2, NOP56, DDX21, SERBP1, SNRPD2, SNRPA1, NME1, ETF1, PSMD8, EEF1B2, PRPF31, CDC45, ERH, GSPT1, SRSF7, MAD2L1, RPLP0, COX5A, SRSF1, CTPS1, DHX15, POLD2, CAD, LSM2, HSPE1, NHP2, ODC1, PSMA1, UBA2, EIF2S1, MRPS18B, TFDP1, RPS2, CCT5, CCNA2, SYNCRIP, HNRNPC, MCM4, PSMD3, RAN, CDC20, RPL22, PSMA4, HSPD1, TCP1, SRSF2, NOP16, PHB2, PHB, NDUFAB1, XRCC6, EIF4E, PCNA, PTGES3, PSMC6, CDK4, PA2G4, PABPC1, PPIA, SRPK1, SRSF3, RPS3, KPNB1, KPNA2, GLO1, TARDBP, C1QBP, SNRPB2, STARD7, ACP1, AIMP2, VDAC3, HNRNPR, PSMC4, RPS6, EIF3B, CCT3, SSBP1, PSMB2, CBX3, RANBP1, AP3S1, CCT2, EIF1AX, G3BP1, EIF2S2, EIF3D, EIF3J, HNRNPA2B1, RPL6, MCM7, CYC1, PSMB3, POLE3, MCM6, HDAC2, VDAC1, SF3A1, CLNS1A, MRPL9, HNRNPD, SLC25A3, XPOT, PSMD7, PPM1G, CANX, PSMD14, TUFM, PRPS2, RPL34, SNRPG, RACK1, CCT7, HNRNPA1, DDX18, SNRPD3, RRM1, PSMA6, DEK, PSMA7 | 3.38 | <1e-44 *** | <1e-42 *** |
| E2F TARGETS | Up | 198 | SMC6, POLD1, MCM2, TFRC, MYC, NOLC1, TRA2B, HELLS, RAD1, MCM5, PAICS, CHEK1, USP1, MTHFD2, LYAR, DUT, POLD3, ZW10, DLGAP5, CCNE1, NOP56, NME1, MLH1, STAG1, TRIP13, SSRP1, TUBG1, GSPT1, BUB1B, PLK4, CDC25A, TCF19, DCTPP1, MAD2L1, PLK1, RAD51AP1, MELK, SRSF1, EXOSC8, CTPS1, DIAPH3, PPP1R8, MYBL2, NASP, POLD2, IPO7, UNG, EIF2S1, KIF2C, RPA1, SYNCRIP, TBRG4, ORC6, MCM4, SMC4, TACC3, RAN, CDC20, JPT1, HMGB2, SRSF2, TOP2A, BRCA1, XRCC6, TIPIN, PCNA, CDK4, MKI67, AURKA, DSCC1, STMN1, PA2G4, GINS4, EED, TUBB, CCP110, WDR90, PSMC3IP, CDCA8, KPNA2, TMPO, MRE11, DCLRE1B, ILF3, KIF22, GINS1, ANP32E, CKS2, NUDT21, HMGA1, SPC25, RANBP1, BRCA2, CENPE, MCM3, UBE2S, E2F8, SPAG5, RAD50, RRM2, GINS3, UBE2T, SMC3, CNOT9, MCM7, RPA2, CKS1B, SLBP, CSE1L, ESPL1, PRKDC, POLE4, MCM6, SUV39H1, MSH2, CDCA3, CDK1, CCNB2, BIRC5, PNN, RAD51C, CTCF, SMC1A, HNRNPD, MMS22L, NUP205, NUP153, POP7 | 3.06 | <1e-30 *** | <1e-28 *** |
| G2M CHECKPOINT | Up | 195 | UCK2, BUB3, MCM2, PRMT5, MYC, NOLC1, TRA2B, AMD1, POLQ, MCM5, CHEK1, SNRPD1, KIF23, CUL4A, SMARCC1, KIF15, SLC38A1, PRC1, STAG1, CDC45, BCL3, NDC80, GSPT1, PLK4, CDC6, CDC25A, MAD2L1, DTYMK, PLK1, NUP50, SRSF1, CCNF, MYBL2, NASP, RAD54L, EWSR1, DKC1, ABL1, EXO1, TTK, ODC1, CENPA, KATNA1, KIF2C, TFDP1, CCNA2, SYNCRIP, ORC6, SMC4, TACC3, CDC20, JPT1, SLC7A1, SRSF2, TOP2A, TRAIP, NCL, CDK4, MKI67, MARCKS, HSPA8, AURKA, STMN1, NSD2, KPNB1, KPNA2, TMPO, SRSF10, FANCC, KIF11, CDC7, ILF3, KIF22, CKS2, INCENP, HMGA1, LIG3, FBXO5, BRCA2, CENPE, MCM3, UBE2S, G3BP1, E2F4, SMC2, TOP1, NUSAP1, RPA2, CKS1B, ESPL1, MCM6, SFPQ, SUV39H1, KIF20B, CDK1, CCNB2, BIRC5, KMT5A, CTCF, SMC1A, RBL1, HNRNPD, ATF5, STIL, HMGN2, E2F2, HIRA, SS18, RBM14, TNPO2, POLA2, CUL5, SLC7A5, HMGB3, MEIS2, MAP3K20 | 2.61 | <1e-17 *** | <1e-16 *** |
| MYC TARGETS V2 | Up | 58 | GNL3, RRP9, IPO4, PPRC1, SRM, RRP12, MYC, NOLC1, MCM5, EXOSC5, GRWD1, NPM1, PES1, SUPV3L1, FARSA, UTP20, PUS1, NOP56, MRTO4, WDR74, PLK4, WDR43, DCTPP1, PLK1, TMEM97, NOP2, BYSL, NOC4L, HSPE1, UNG, RABEPK, TBRG4, MCM4, TCOF1, HSPD1, NOP16, PHB, MYBBP1A, CDK4, PRMT3, SLC19A1, PA2G4, AIMP2, TFB2M, CBX3 | 2.91 | <1e-15 *** | <1e-14 *** |
| OXIDATIVE PHOSPHORYLATION | Up | 200 | NDUFV2, TOMM70, ATP5F1A, IDH3A, TIMM50, DLAT, GOT2, NDUFB4, MRPS22, OPA1, LDHB, SUPV3L1, MRPL11, NDUFA9, ECI1, OXA1L, PDHB, ACAA2, FH, PMPCA, ATP5MC1, COX5A, ETFA, SUCLA2, LRPPRC, MRPS12, AFG3L2, MRPS11, BAX, UQCRH, ALDH6A1, GLUD1, CYCS, HSPA9, COX6B1, UQCRFS1, ACADM, TIMM17A, ATP5F1D, PHB2, NDUFAB1, NDUFB5, IMMT, MRPS15, ACAA1, TIMM13, GPI, ATP5MC2, NDUFS2, GRPEL1, UQCRC1, VDAC3, NDUFA3, MTRR, OGDH, NDUFS3, HCCS, SDHC, TOMM22, TIMM10, ECHS1, TIMM8B, MRPS30, MRPL15, POLR2F, MTX2, ACADSB, ECH1, CYC1, VDAC1, COX17, NDUFS4, NDUFC2, ATP1B1, CASP7, SLC25A3, ACAT1, MDH2, ATP5PD, COX11, TIMM9, VDAC2, DECR1, SLC25A12, NDUFB8, ATP5ME, ATP5F1E, UQCRQ, SDHA, NDUFS8, HTRA2, NDUFB7, AIFM1, UQCR11, COX7C, COX15, NDUFV1, FXN, SDHB, COX10, ATP5PB, PDHX, SLC25A6, ATP5F1B, ETFDH, PRDX3, NDUFS6, UQCRC2, NDUFB6, RHOT2, CPT1A, ISCA1, COX4I1, MRPL35, HADHA, IDH3B, HADHB, SDHD, COX8A, NDUFA8 | 2.47 | <1e-14 *** | <1e-13 *** |
| DNA REPAIR | Up | 147 | PDE4B, RFC4, ELOA, POLR2I, POLR1C, POLD1, SEC61A1, PNP, POLR2H, ERCC2, TYMS, CSTF3, NCBP2, UMPS, DUT, POLD3, IMPDH2, POLR1D, NFX1, SUPT5H, MRPL40, NME1, SSRP1, NT5C, ZWINT, SNAPC4, EIF1B, ERCC1, MPG, RNMT, NME4, PCNA, POLR2E, ELL, RAD51, SRSF6, EDF1, RFC5, FEN1, ARL6IP1, NELFCD, NUDT21, ALYREF, SF3A3, APRT, DAD1, POLR2D, POLR3C, PRIM1, POLR2F, DDB1, POLB, GTF3C5, GSDME, RPA2, ERCC8, POLE4, POLR2C, COX17, GTF2B, TAF9, GTF2A2, AAAS, GTF2H3, LIG1, GTF2H1, POLA2, NELFB, NELFE, ADA | 2.24 | <1e-09 *** | <1e-08 *** |
| MTORC1 SIGNALING | Up | 197 | SLA, RRP9, HSP90B1, POLR3G, GCLC, GMPS, MCM2, TFRC, TOMM40, PNP, MTHFD2L, MTHFD2, RPN1, SERP1, TRIB3, NFKBIB, PSMA3, ETF1, CYB5B, USO1, TUBG1, CDC25A, UCHL5, PLK1, HMBS, ATP5MC1, TMEM97, CCNF, HSPE1, NUFIP1, ARPC5L, UNG, SLC1A5, HSPA9, RPA1, XBP1, MCM4, PSMG1, PSMA4, HSPD1, IMMT, PSMC6, QDPR, PSMB5, NMT1, AURKA, PPIA, INSIG1, GPI, PSMC4, PSMD13, UFM1, BTG2, CTH, CXCR4, PSME3, ABCF2, ACLY, SHMT2, ENO1, HK2, PSMD12, EIF2S2, EEF1E1, RRM2, PNO1, CACYBP, SSR1, NFYC, CCT6A, STIP1, CALR, SEC11A, UBE2D3, STARD4, CORO1A, CTSC, NAMPT, PSAT1, NUP205, GSK3B, CANX, PITPNB, PSMD14, GTF2H1, PPA1, SLC7A5 | 2.12 | <1e-08 *** | <1e-08 *** |
| UNFOLDED PROTEIN RESPONSE | Up | 108 | RRP9, HSP90B1, EIF4G1, NOLC1, EXOSC5, MTHFD2, SDAD1, SERP1, NPM1, CXXC1, SPCS1, NOP14, NOP56, SRPRB, DCP1A, EIF4A2, EXOSC2, ALDH18A1, DDX10, EIF4A3, DNAJA4, DKC1, ZBTB17, NHP2, ERN1, EIF2S1, HSPA9, XBP1, EXOSC9, SPCS3, EIF4E, EXOSC10, FUS, BANF1, POP4, TATDN2, GEMIN4, EXOSC1, MTREX, YIF1A, NABP1, ATF4, SEC31A, CKS1B, SSR1, HYOU1, CALR, SEC11A, EDC4, IMP3, GOSR2, PDIA5, PSAT1, KHSRP, XPOT, RPS14, SLC7A5, ATF3, PDIA6, ARFGAP1, EXOSC4 | 2.26 | <1e-08 *** | <1e-07 *** |
| FATTY ACID METABOLISM | Up | 146 | EHHADH, ERP29, APEX1, GCDH, RAP1GDS1, METAP1, SUCLG2, ACSM3, ENO3, ECI1, GRHPR, UBE2L6, PDHB, ACAA2, CPOX, GPD2, FH, SUCLA2, ODC1, CRYZ, ACADM, HSPH1, ACAA1, OSTC, PSME1, MIF, HCCS, ADSL, SDHC, HADH, ECHS1, BCKDHB, CD36, SMS, ECH1, ALDH3A1, CD1D, NTHL1, HSD17B4, KMT5A, HSD17B11, ECI2, HSP90AA1, MDH2, DECR1, SDHA, ALDH3A2, ACADS, PCBD1, MCEE | 1.49 | 0.0055 ** | 0.0124 * |
| REACTIVE OXYGEN SPECIES PATHWAY | Up | 48 | PDLIM1, GCLC, OXSR1, ERCC2, NDUFB4, PFKP, MGST1, HMOX2, NDUFS2, MBP, PTPA, PRDX2, TXN, CDKN2D, STK25, GSR, HHEX, PRDX4, GLRX2, SBNO2 | 1.53 | 0.0200 * | 0.0385 * |
| GenesetID | Direction | Size | Leading.Genes | NES | P.Value | adj.P.Val |
|---|---|---|---|---|---|---|
| EPITHELIAL MESENCHYMAL TRANSITION | Down | 194 | PMEPA1, ITGAV, MXRA5, ADAM12, FAP, BGN, DPYSL3, MMP2, CDH11, DCN, POSTN, FBN1, LRRC15, TPM1, LUM, COL16A1, SGCD, ECM2, COL5A1, FN1, COL1A2, PLOD2, THBS2, COL6A3, CALD1, COL12A1, COL3A1, COL1A1, COL5A2, FSTL1, TFPI2, BMP1, FBLN2, GLIPR1, PCOLCE, INHBA, SPARC, LAMA2, HTRA1, FZD8, CTHRC1, SNAI2, MMP14, ITGB5, VCAN, NNMT, DST, EDIL3, COL6A2, FERMT2, ECM1, LRP1, TIMP3, SPOCK1, FBLN5, FBLN1, WIPF1, PMP22, COL11A1, MYLK, PDGFRB, THY1, WNT5A, FSTL3, MSX1, CDH6, PRRX1, IGFBP2, SERPINH1, PDLIM4, MYL9, GPC1, NID2, GAS1, LOXL1, ITGA5, DAB2, LAMA1, EFEMP2, SGCB, ACTA2, MATN3, SDC4, MGP, PLAUR, ITGB3, ELN, COL7A1, SLIT2, CAP2, PLOD1, IGFBP4, BDNF, COL4A2, GEM, TGFBI, TAGLN, LOX, ABI3BP, COL5A3, FMOD, SDC1, FGF2, SERPINE2, PVR, TIMP1, P3H1, COL4A1, ANPEP, CALU, MMP3, NT5E, GJA1, THBS1, RHOB, CAPG, COMP, SNTB1, QSOX1, SFRP4, MATN2, CXCL6, ITGB1, APLP1, FAS, CXCL8, SLIT3, RGS4, CRLF1, MAGEE1 | -3.18 | <1e-31 *** | <1e-29 *** |
| MYOGENESIS | Down | 176 | ABLIM1, ADAM12, SGCD, PLXNB2, SSPN, COL6A3, COL3A1, COL1A1, SPARC, NAV2, LAMA2, ACTN2, SVIL, MYH9, AEBP1, ITGB5, COL6A2, MYH11, CHRNA1, CNN3, MEF2C, BDKRB2, WWTR1, MYLK, PICK1, EPHB3, GSN, PRNP, ADCY9, PPFIA4, CACNA1H, MYOM2, AGRN, MYO1C, IGFBP7, MYBPC3, SGCA, EFS, CAMK2B, SPHK1, COL15A1, PSEN2, DMPK, HSPB8, PTGIS, REEP1, CRYAB, CDH13, NOS1, LSP1, FXYD1, COL4A2, TAGLN, HSPB2, KIFC3, PDLIM7, TNNT2, APLNR, FGF2, PC, SOD3, DMD, NCAM1, GABARAPL2, SPTAN1, SPEG, TPD52L1, LDB3, ERBB3, PPP1R3C, GJA5, COX7A1, DTNA, MYL2, PDE4DIP, MRAS, IGF1, LPIN1, OCEL1, SORBS1, SYNGR2, CASQ2, ITGB1, TNNT3, PGAM2, HBEGF, DAPK2, PYGM, CASQ1, IGFBP3, TEAD4, FHL1, IFRD1, HRC, CRAT, CKMT2, MYH8 | -2.64 | <1e-16 *** | <1e-15 *** |
| COAGULATION | Down | 114 | KLF7, SH2B2, MMP2, FBN1, CPQ, FN1, TFPI2, BMP1, CTSK, SPARC, MSRB2, OLR1, HTRA1, MMP14, C1S, PLAT, PRSS23, C3, CD9, LRP1, F3, TIMP3, PLAU, MMP7, GSN, C1R, GNG12, MMP9, MMP11, MEP1A, ITIH1, ACOX2, CFI, ITGB3, ANXA1, CFH, TIMP1, LAMP2, MAFF, MMP8, USP11, MMP3, F2RL2, THBS1, RAC1, COMP, PROS1, PDGFB, CSRP1, APOC1, CTSO | -2.43 | <1e-09 *** | <1e-09 *** |
| UV RESPONSE DN | Down | 142 | LTBP1, SYNE1, APBB2, SMAD7, GCNT1, NR3C1, COL1A2, COL3A1, COL1A1, COL5A2, PTPN21, ATXN1, ICA1, PRKCA, CACNA1A, SNAI2, PTPRM, CDK13, AMPH, TJP1, ATP2B1, F3, SYNJ2, FBLN5, MAGI2, PMP22, COL11A1, PLPP3, PDGFRB, KCNMA1, ADD3, DLC1, IRS1, SDC2, ERBB2, MAPK14, SMAD3, MGMT, DAB2, MAP1B, FHL2, ACVR2A, ITGB3, ANXA4, CDON, CAP2, MMP16, EFEMP1, BDNF, KIT, LPAR1, IGFBP5, GRK5, RBPMS, NRP1, GJA1, SLC22A18, IGF1R, ATP2B4, NFIB, WDR37, HAS2, PTGFR, VLDLR, RGS4, LDLR, RND3, ID1, ADORA2B, PRKAR2B, CDC42BPA, AKT3, PDLIM5, NOTCH2, ADGRL2, SCAF8, PLCB4, ATRN, KALRN | -2.26 | <1e-08 *** | <1e-08 *** |
| APICAL JUNCTION | Down | 183 | PTK2, VCL, EPB41L2, MMP2, CDH11, FBN1, COL16A1, EVL, PARVA, NF1, NLGN2, BMP1, EGFR, CERCAM, ACTN2, ACTN1, MYH9, JUP, SHC1, VCAN, TJP1, NECTIN3, FSCN1, PLCG1, WASL, LIMA1, THY1, CTNND1, THBS3, CDH6, LAMB3, IRS1, ADAM23, SHROOM2, MMP9, MYL9, MAPK14, MAPK13, CD276, ADAMTS5, CD86, SLIT2, TGFBI, GNAI1, GAMT, SIRPA, MSN, FLNC, BAIAP2, TRAF1, RSU1, CD99, JAM3, PCDH1, RHOF, SPEG, NEXN, STX4, ATP1A3, COL9A1, NEGR1, ICAM1, ALOX15B, AMH, ITGB1, TSC1, RRAS, CDH8, PTPRC, MDK, ZYX, CDH1, NLGN3, CDH3, NECTIN4, LAMC2, NFASC, SRC, CRAT, AKT3, DSC3, MAP4K2, MYH10, FYB1, EXOC4, LAYN, CTNNA1, TNFRSF11B, PKD1, LDLRAP1 | -2.10 | <1e-08 *** | <1e-07 *** |
| HYPOXIA | Down | 182 | KLF7, BGN, DCN, PDK3, KLHL24, COL5A1, CCNG2, EXT1, NR3C1, PFKL, KDM3A, TPBG, EGFR, PRKCA, MYH9, PFKFB3, P4HA2, ATP7A, GPC4, TPD52, F3, DDIT4, SLC6A6, TMEM45A, BNIP3L, ACKR3, PPFIA4, PDK1, ERRFI1, SDC2, GPC1, CHST3, P4HA1, DDIT3, PCK1, LXN, IDS, HAS1, KDELR3, PPARGC1A, SDC4, PLAUR, NFIL3, SELENBP1, FOSL2, PLIN2, TGFBI, LOX, GCNT2, MAFF, FBP1, FOXO3, CA12, AMPD3, PPP1R3C, PAM, DTNA, SAP30, CSRP2, TGFB3, TPST2, UGP2, EFNA1, CAVIN1, SLC25A1, PDGFB, RRAGD, PGAM2, SRPX, VLDLR, IER3, PYGM, PKP1, PNRC1, IGFBP3, ADORA2B, DPYSL4, NDRG1, FOS, RORA, LARGE1, ALDOB, SLC2A3, TES, AKAP12 | -1.99 | <1e-07 *** | <1e-06 *** |
| ANGIOGENESIS | Down | 32 | PTK2, ITGAV, POSTN, LUM, COL3A1, COL5A2, FSTL1, OLR1, FGFR1, VCAN, JAG1, JAG2, MSX1 | -2.00 | 0.0004 *** | 0.0012 ** |
| XENOBIOTIC METABOLISM | Down | 171 | ALDH2, TPST1, SLC35D1, UPP1, IL1R1, SLC12A4, ABCC3, JUP, DHRS7, GABARAPL1, LCAT, DHRS1, FBLN1, BCAR1, SLC6A6, PAPSS2, CSAD, PINK1, ESR1, CYP2S1, BPHL, CES1, TGFB2, CYP27A1, PTGR1, ELOVL5, KYNU, ITIH1, ACOX2, MAOA, CYP2E1, CROT, IGFBP4, ACO2, PTS, ACOX1, TNFRSF1A, FMO1, GCNT2, PC, CDA, CYB5A, AKR1C3, TDO2, FBP1, ENPEP, PTGES, PROS1, GAD1, IGF1, NDRG2, UPB1, ETS2, POR, CYP1A1, APOE | -1.56 | 0.0008 *** | 0.0026 ** |
| INFLAMMATORY RESPONSE | Down | 188 | LCK, SLAMF1, IL1R1, RGS16, PTAFR, TPBG, INHBA, PTGIR, OLR1, PCDH7, MMP14, ACVR1B, ATP2B1, IRAK2, F3, PDPN, GPR132, SCN1B, DCBLD2, ROS1, SPHK1, ITGA5, P2RX4, MEP1A, TLR1, ACVR2A, TNFAIP6, PLAUR, ITGB3, AXL, ITGB8, CSF1, HRH1, ABCA1, LPAR1, SLC4A4, RGS1, LAMP3, HIF1A, LCP2, CLEC5A, APLNR, TLR3, CALCRL, NLRP3, PVR, RIPK2, TIMP1, IL4R, SGMS2, BST2, IL18, BDKRB1, SLC7A2, EIF2AK2, ICAM1, BEST1, CXCL6, SELE, HAS2, MXD1, NDP, LIF, HBEGF, CXCL8, LDLR, CCL17, OSMR, SLC1A2, ADORA2B, TLR2, SLC11A2, TNFRSF9, OSM, IL15, CSF3R, EMP3, PSEN1, IL7R, CYBB | -1.53 | 0.0019 ** | 0.0055 ** |
| KRAS SIGNALING UP | Down | 181 | SDCCAG8, RGS16, ANO1, ADAM17, ADGRA2, INHBA, GUCY1A1, JUP, KIF5C, ITGBL1, GFPT2, ALDH1A3, PLAT, SPON1, TRIB2, PLAU, PEG3, PRKG2, SOX9, USP12, PRRX1, CSF2RA, MMP9, GADD45G, MMP11, SCN1B, DCBLD2, CAB39L, SATB1, EMP1, ABCB1, IL1RL2, SPRY2, PLEK2, SEMA3B, PLAUR, CFH, CROT, CPE, CLEC4A, ITGB2, BMP2, TNNT2, SERPINA3, TSPAN1, TRAF1, VWA5A, NRP1, BTBD3, MAP7, SNAP25, F2RL1, GALNT3, GNG11, LIF, HBEGF, ALDH1A2, G0S2, ENG, LAT2, DOCK2, EVI5, IGFBP3, HDAC9, ETV1, PTGS2 | -1.49 | 0.0030 ** | 0.0080 ** |
| ESTROGEN RESPONSE EARLY | Down | 185 | MYBL1, ASB13, SLC1A1, ABLIM1, FAM102A, SLC24A3, PDLIM3, RETREG1, TPBG, NAV2, SEC14L2, SVIL, NCOR2, NRIP1, SYBU, KLF10, PRSS23, TSKU, AR, PAPSS2, ADD3, ADCY9, ELOVL2, DLC1, FLNB, OLFML3, FHL2, CCND1, PODXL, ELOVL5, HSPB8, SEMA3B, REEP1, DEPTOR, TFAP2C, CLIC3, IGFBP4, ELF1, DYNLT3, TOB1, SLC37A1, TIAM1, LAD1, ZNF185, MLPH, DHRS3, TPD52L1, CA12, PGR, GREB1, GJA1, ABHD2, IGF1R, SLC7A2, MED13L, HR, PTGES, MAST4, TGIF2, GFRA1, SCNN1A, SYNGR1, THSD4, KRT19 | -1.51 | 0.0030 ** | 0.0080 ** |
| TNFA SIGNALING VIA NFKB | Down | 196 | BCL6, PMEPA1, RNF19B, SPSB1, REL, SGK1, IER2, CLCF1, INHBA, B4GALT5, OLR1, GFPT2, KLF10, PFKFB3, ATP2B1, F3, PLAU, JAG1, DNAJB4, PLPP3, PLK2, ACKR3, LAMB3, DENND5A, SMAD3, MSC, SPHK1, CCND1, MAP3K8, TNFAIP6, KYNU, IFIH1, SDC4, BIRC2, PLAUR, DUSP4, NFIL3, FOSL2, CSF1, GEM, ABCA1, BMP2, SLC2A6, RIPK2, TRAF1, TNIP2, MAFF, TSC22D1, ZBTB10, RCAN1, IL18, RHOB, EFNA1, TRIP10, PHLDA2, ETS2, ICAM1, F2RL1, CXCL6, NFE2L2, EGR1, MXD1, TUBB2A, SQSTM1, LIF, HBEGF, G0S2, IER3, LDLR, CD83, TNFAIP2, PNRC1, GADD45A, TLR2, PTGS2, TNFRSF9, JUNB, PDLIM5, FOS, SLC2A3, IL7R, EGR2, CEBPB, TNC, BIRC3, KDM6B, KLF4, KLF9, SLC16A6, NFKB2, CEBPD, DUSP1, IL1A | -1.44 | 0.0047 ** | 0.0116 * |
| TGF BETA SIGNALING | Down | 52 | PMEPA1, SMAD7, LTBP2, HDAC1, SMAD1, NCOR2, SMURF2, KLF10, TJP1, WWTR1, SMURF1, SMAD3, TGFBR1, SMAD6, ACVR1, PPM1A, BMP2 | -1.62 | 0.0053 ** | 0.0124 * |
| HEDGEHOG SIGNALING | Down | 32 | NF1, MYH9, DPYSL2, NRP2, THY1, OPHN1, RTN1, HEY2, UNC5C, NRP1, HEY1, ETS2, CRMP1, VLDLR, AMOT, CELSR1 | -1.69 | 0.0091 ** | 0.0198 * |
| NOTCH SIGNALING | Down | 32 | WNT2, PRKCA, JAG1, NOTCH3, WNT5A, ST3GAL6, HEYL, FZD1, PSEN2, CCND1, TCF7L2, DTX1, DTX2 | -1.67 | 0.0106 * | 0.0220 * |
| APICAL SURFACE | Down | 40 | EFNA5, CRYBG1, AKAP7, THY1, PKHD1, SHROOM2, DCBLD2, GAS1, GHRL, PLAUR, GSTM3, HSPB1, AFAP1L2, LYPD3, NTNG1, CROCC, ATP8B1, SRPX, MDGA1, GATA3, SCUBE1 | -1.56 | 0.0169 * | 0.0337 * |
names(significant_gsea_list) <- contrastsGSEA summary
############################################################################# GSEA
############################################################################# summary
significant_gsea <- unique(unlist(significant_gsea_list))
topTable_significant_gsea <- topTable_gsea[significant_gsea, , drop = FALSE]
data_lfc <- wrapper_extract_from_topTable(topTable_significant_gsea, extract_prefix = "NES")
if (ncol(data_lfc) > 1) {
# Cluster rows using hierarchical clustering
d <- dist(data_lfc, method = "euclidean")
cluster_rows <- hclust(d, method = "complete")
clustering_order <- cluster_rows$order
} else {
cluster_rows <- FALSE
clustering_order <- 1:nrow(data_lfc)
}
### logFC_dotplot
wrapper_logFC_dotplot(topTable_significant_gsea[clustering_order, ], gene_var = "GenesetID",
lfc_prefix = "NES", pval_prefix = "P.Value", adjp_prefix = "adj.P.Val", axis_text_y_size = 10,
trim_values = 4, pval = pval, title = "Gene Set Enrichment Analysis")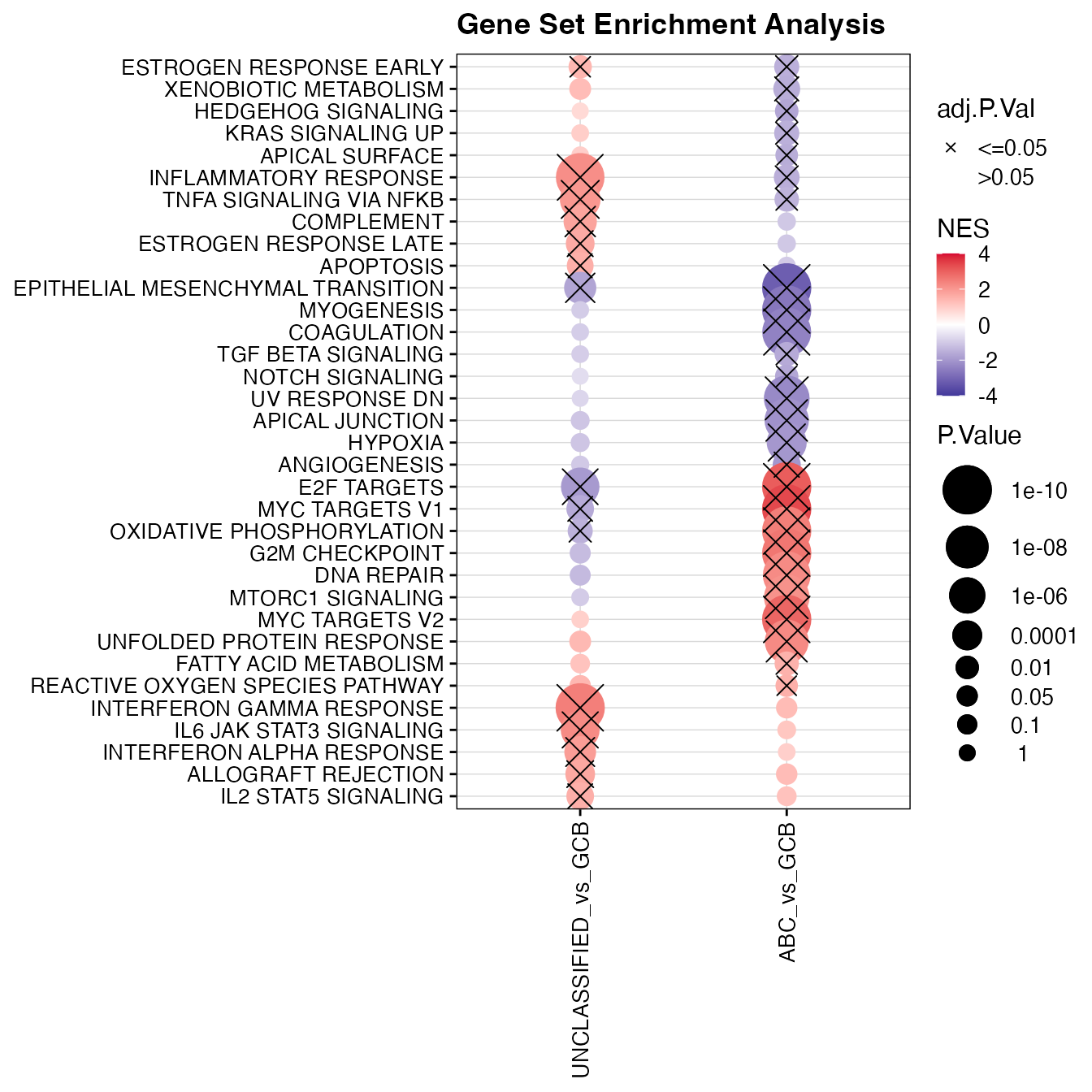
### logFC_heatmap
wrapper_logFC_heatmap(topTable_significant_gsea, gene_var = "GenesetID", lfc_prefix = "NES",
adjp_prefix = "adj.P.Val", cluster_rows = cluster_rows, row_names_gp = grid::gpar(fontsize = 8,
lineheight = 0.8), trim_values = 4)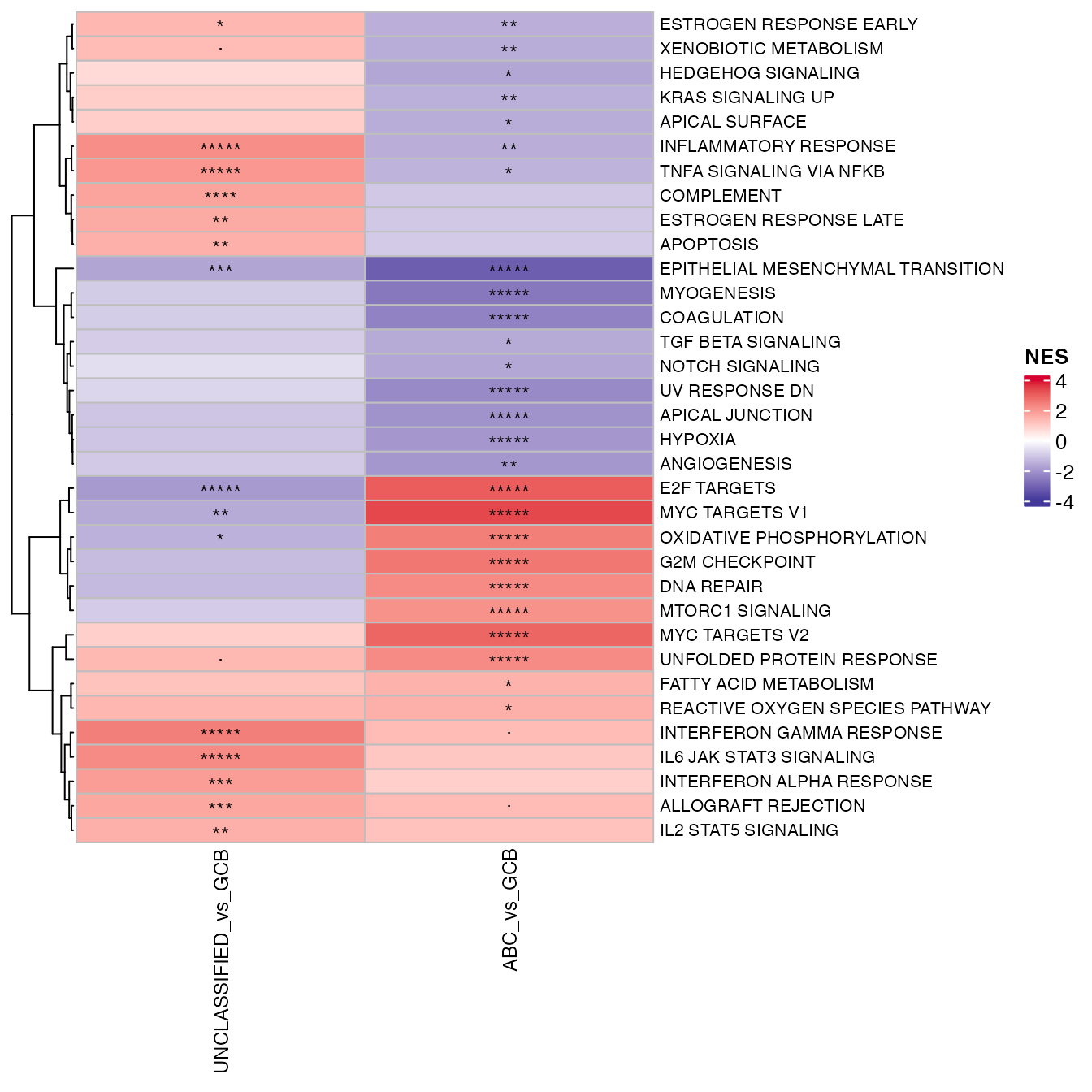
## R version 4.4.1 (2024-06-14)
## Platform: x86_64-apple-darwin20
## Running under: macOS Sonoma 14.6.1
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: Europe/Zurich
## tzcode source: internal
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] BiomarkerWrappers_0.0.0.60 msigdbr_7.5.1
## [3] edgeR_4.2.0 limma_3.60.3
## [5] SummarizedExperiment_1.34.0 Biobase_2.64.0
## [7] GenomicRanges_1.56.1 GenomeInfoDb_1.40.1
## [9] IRanges_2.38.0 S4Vectors_0.42.0
## [11] BiocGenerics_0.50.0 MatrixGenerics_1.16.0
## [13] matrixStats_1.3.0 ggplot2_3.5.1
## [15] kableExtra_1.4.0 rmarkdown_2.27
## [17] knitr_1.47
##
## loaded via a namespace (and not attached):
## [1] polynom_1.4-1 formatR_1.14 rlang_1.1.4
## [4] magrittr_2.0.3 clue_0.3-65 GetoptLong_1.0.5
## [7] compiler_4.4.1 png_0.1-8 systemfonts_1.1.0
## [10] vctrs_0.6.5 stringr_1.5.1 pkgconfig_2.0.3
## [13] shape_1.4.6.1 crayon_1.5.3 fastmap_1.2.0
## [16] magick_2.8.3 XVector_0.44.0 labeling_0.4.3
## [19] utf8_1.2.4 UCSC.utils_1.0.0 ragg_1.3.2
## [22] purrr_1.0.2 xfun_0.45 zlibbioc_1.50.0
## [25] cachem_1.1.0 jsonlite_1.8.8 highr_0.11
## [28] DelayedArray_0.30.1 BiocParallel_1.38.0 parallel_4.4.1
## [31] cluster_2.1.6 R6_2.5.1 bslib_0.7.0
## [34] stringi_1.8.4 RColorBrewer_1.1-3 jquerylib_0.1.4
## [37] Rcpp_1.0.12 iterators_1.0.14 Matrix_1.7-0
## [40] splines_4.4.1 tidyselect_1.2.1 rstudioapi_0.16.0
## [43] abind_1.4-5 yaml_2.3.8 doParallel_1.0.17
## [46] codetools_0.2-20 plyr_1.8.9 lattice_0.22-6
## [49] tibble_3.2.1 withr_3.0.0 evaluate_0.24.0
## [52] desc_1.4.3 survival_3.7-0 ggpp_0.5.8
## [55] xml2_1.3.6 circlize_0.4.16 pillar_1.9.0
## [58] corrplot_0.92 foreach_1.5.2 generics_0.1.3
## [61] munsell_0.5.1 scales_1.3.0 glue_1.7.0
## [64] tools_4.4.1 data.table_1.15.4 fgsea_1.30.0
## [67] locfit_1.5-9.10 babelgene_22.9 fs_1.6.4
## [70] fastmatch_1.1-4 Cairo_1.6-2 cowplot_1.1.3
## [73] grid_4.4.1 tidyr_1.3.1 colorspace_2.1-0
## [76] GenomeInfoDbData_1.2.12 cli_3.6.3 textshaping_0.4.0
## [79] fansi_1.0.6 S4Arrays_1.4.1 viridisLite_0.4.2
## [82] svglite_2.1.3 ComplexHeatmap_2.20.0 dplyr_1.1.4
## [85] gtable_0.3.5 sass_0.4.9 digest_0.6.36
## [88] ggrepel_0.9.5 SparseArray_1.4.8 farver_2.1.2
## [91] rjson_0.2.21 htmlwidgets_1.6.4 memoise_2.0.1
## [94] htmltools_0.5.8.1 pkgdown_2.0.9 lifecycle_1.0.4
## [97] httr_1.4.7 GlobalOptions_0.1.2 statmod_1.5.0
## [100] MASS_7.3-61